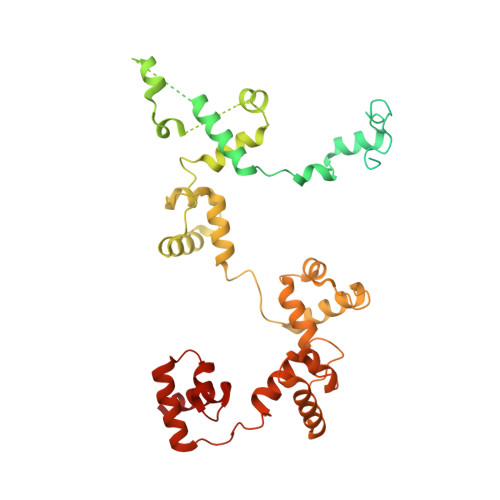
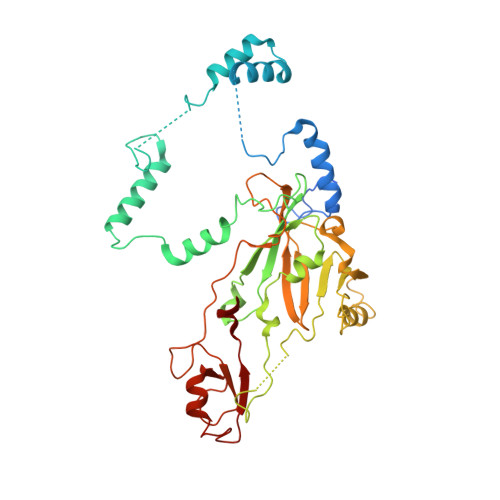
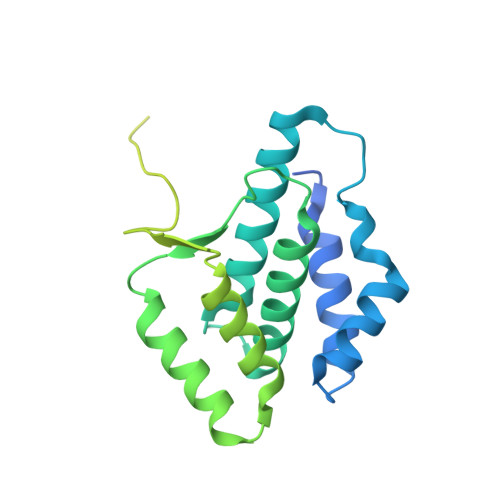
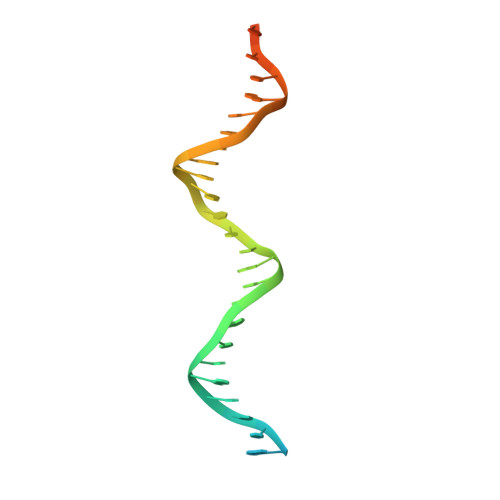
Structural basis of human SNAPc recognizing proximal sequence element of snRNA promoter.
Sun, J., Li, X., Hou, X., Cao, S., Cao, W., Zhang, Y., Song, J., Wang, M., Wang, H., Yan, X., Li, Z., Roeder, R.G., Wang, W.(2022) Nat Commun 13: 6871-6871
- PubMed: 36369505
- DOI: https://doi.org/10.1038/s41467-022-34639-1
- Primary Citation of Related Structures:
7XUR - PubMed Abstract:
In eukaryotes, small nuclear RNAs (snRNAs) function in many fundamental cellular events such as precursor messenger RNA splicing, gene expression regulation, and ribosomal RNA processing. The snRNA activating protein complex (SNAPc) exclusively recognizes the proximal sequence element (PSE) at snRNA promoters and recruits RNA polymerase II or III to initiate transcription. In view that homozygous gene-knockout of SNAPc core subunits causes mouse embryonic lethality, functions of SNAPc are almost housekeeping. But so far, the structural insight into how SNAPc assembles and regulates snRNA transcription initiation remains unclear. Here we present the cryo-electron microscopy structure of the essential part of human SNAPc in complex with human U6-1 PSE at an overall resolution of 3.49 Å. This structure reveals the three-dimensional features of three conserved subunits (N-terminal domain of SNAP190, SNAP50, and SNAP43) and explains how they are assembled into a stable mini-SNAPc in PSE-binding state with a "wrap-around" mode. We identify three important motifs of SNAP50 that are involved in both major groove and minor groove recognition of PSE, in coordination with the Myb domain of SNAP190. Our findings further elaborate human PSE sequence conservation and compatibility for SNAPc recognition, providing a clear framework of snRNA transcription initiation, especially the U6 system.
- Advanced Medical Research Institute, Cheeloo College of Medicine, Shandong University, Jinan, 250012, China. sunjf@sdu.edu.cn.
Organizational Affiliation: