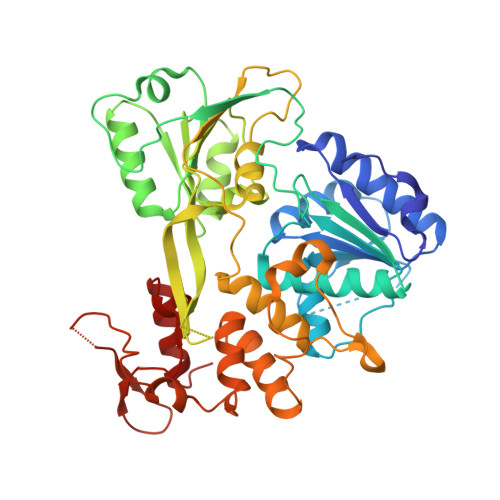
Crystal structure of RNA helicase from Saint Louis encephalitis virus and discovery of its inhibitors.
Wang, D.P., Jiang, F.Y., Zeng, X.Y., Liu, Y.J., Zhao, R., Wang, M.Y., Luo, J., Chen, C., Zhu, Y., Cao, J.M.(2023) Genes Dis 10: 389-392
- PubMed: 37223502
- DOI: https://doi.org/10.1016/j.gendis.2022.07.001
- Primary Citation of Related Structures:
7XT0 - Key Laboratory of Cellular Physiology at Shanxi Medical University, Ministry of Education, And the Department of Physiology, Shanxi Medical University, Taiyuan, Shanxi 030001, China.
Organizational Affiliation: