Enzymatic conversion of CO2 in real flue gas to molar-scale formate
Jeon, B.W., Heo, Y.Y., Park, J., Roh, S.H., Lee, H.H., Kim, Y.H.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Formate dehydrogenase | 995 | Methylorubrum extorquens AM1 | Mutation(s): 0 Gene Names: fdh1A, MexAM1_META1p5032 EC: 1.17.1.9 (PDB Primary Data), 1.2.1.2 (UniProt) | 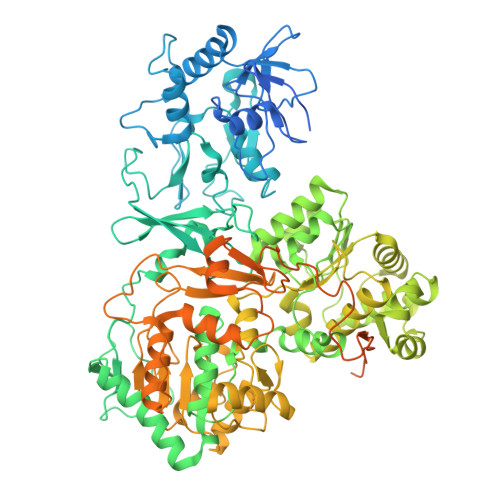 | |
UniProt | |||||
Find proteins for C5ATT7 (Methylorubrum extorquens (strain ATCC 14718 / DSM 1338 / JCM 2805 / NCIMB 9133 / AM1)) Explore C5ATT7 Go to UniProtKB: C5ATT7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | C5ATT7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Tungsten-containing formate dehydrogenase beta subunit | 572 | Methylorubrum extorquens AM1 | Mutation(s): 0 Gene Names: fdh1B, MexAM1_META1p5031 EC: 1.2.1.2 | 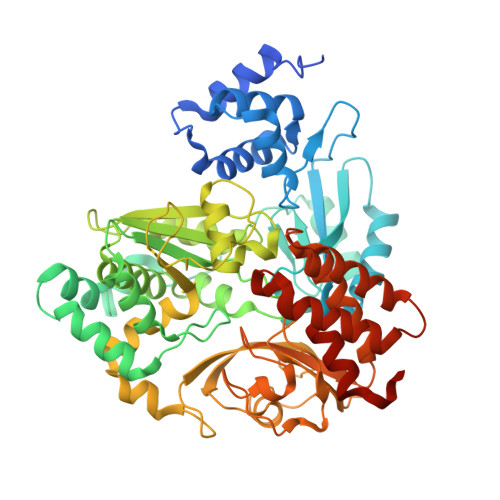 | |
UniProt | |||||
Find proteins for C5ATT6 (Methylorubrum extorquens (strain ATCC 14718 / DSM 1338 / JCM 2805 / NCIMB 9133 / AM1)) Explore C5ATT6 Go to UniProtKB: C5ATT6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | C5ATT6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 5 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| MGD (Subject of Investigation/LOI) Query on MGD | D [auth A], E [auth A] | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE C20 H26 N10 O13 P2 S2 VQAGYJCYOLHZDH-ILXWUORBSA-N |  | ||
| FMN (Subject of Investigation/LOI) Query on FMN | J [auth B] | FLAVIN MONONUCLEOTIDE C17 H21 N4 O9 P FVTCRASFADXXNN-SCRDCRAPSA-N |  | ||
| SF4 Query on SF4 | G [auth A], H [auth A], I [auth A], K [auth B] | IRON/SULFUR CLUSTER Fe4 S4 LJBDFODJNLIPKO-UHFFFAOYSA-N |  | ||
| W Query on W | C [auth A] | TUNGSTEN ION W FZFRVZDLZISPFJ-UHFFFAOYSA-N |  | ||
| FES Query on FES | F [auth A], L [auth B] | FE2/S2 (INORGANIC) CLUSTER Fe2 S2 NIXDOXVAJZFRNF-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| RECONSTRUCTION | cryoSPARC | 3.2 |
| MODEL REFINEMENT | PHENIX | 1.19 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Research Foundation (NRF, Korea) | Korea, Republic Of | 2021M3A9I4021220 |
| National Research Foundation (NRF, Korea) | Korea, Republic Of | 2019M3E5D6063871 |