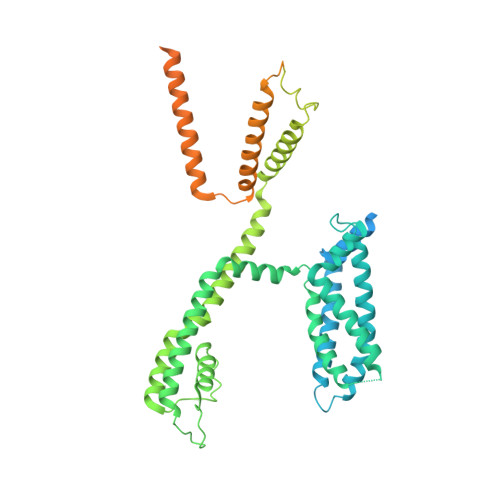
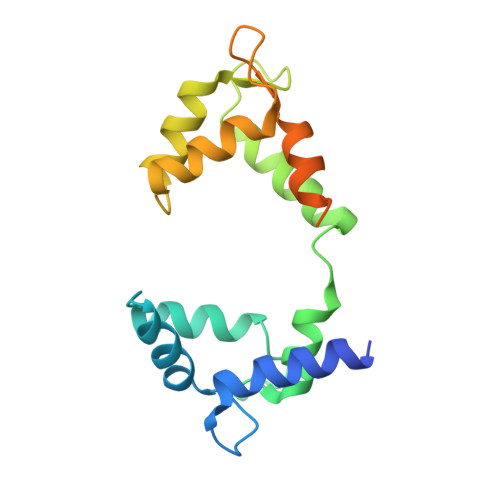
Structural mechanisms for the activation of human cardiac KCNQ1 channel by electro-mechanical coupling enhancers.
Ma, D., Zhong, L., Yan, Z., Yao, J., Zhang, Y., Ye, F., Huang, Y., Lai, D., Yang, W., Hou, P., Guo, J.(2022) Proc Natl Acad Sci U S A 119: e2207067119-e2207067119
- PubMed: 36763058
- DOI: https://doi.org/10.1073/pnas.2207067119
- Primary Citation of Related Structures:
7XNI, 7XNK, 7XNL, 7XNN - PubMed Abstract:
The cardiac KCNQ1 potassium channel carries the important I Ks current and controls the heart rhythm. Hundreds of mutations in KCNQ1 can cause life-threatening cardiac arrhythmia. Although KCNQ1 structures have been recently resolved, the structural basis for the dynamic electro-mechanical coupling, also known as the voltage sensor domain-pore domain (VSD-PD) coupling, remains largely unknown. In this study, utilizing two VSD-PD coupling enhancers, namely, the membrane lipid phosphatidylinositol 4,5-bisphosphate (PIP 2 ) and a small-molecule ML277, we determined 2.5-3.5 Å resolution cryo-electron microscopy structures of full-length human KCNQ1-calmodulin (CaM) complex in the apo closed, ML277-bound open, and ML277-PIP 2 -bound open states. ML277 binds at the "elbow" pocket above the S4-S5 linker and directly induces an upward movement of the S4-S5 linker and the opening of the activation gate without affecting the C-terminal domain (CTD) of KCNQ1. PIP 2 binds at the cleft between the VSD and the PD and brings a large structural rearrangement of the CTD together with the CaM to activate the PD. These findings not only elucidate the structural basis for the dynamic VSD-PD coupling process during KCNQ1 gating but also pave the way to develop new therapeutics for anti-arrhythmia.
- Department of Biophysics, and Department of Neurology of the Fourth Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, 310058, China.
Organizational Affiliation: