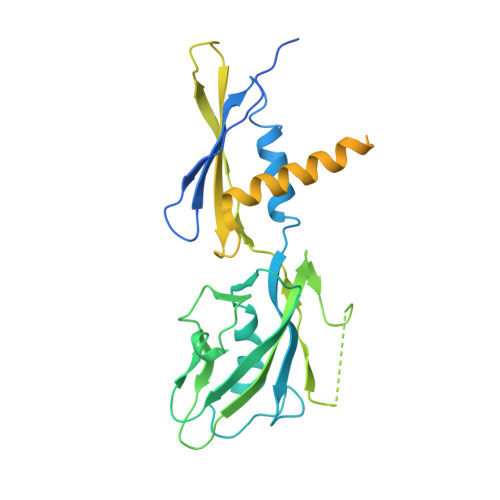
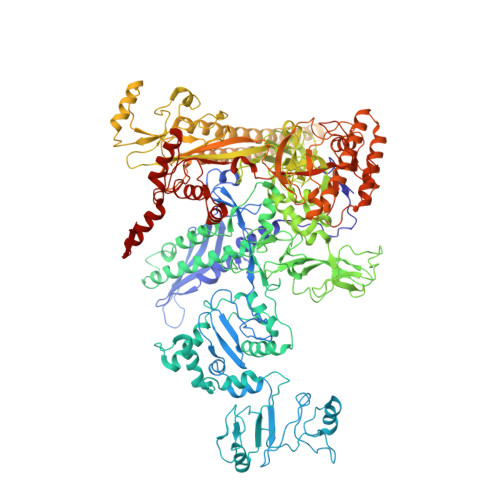
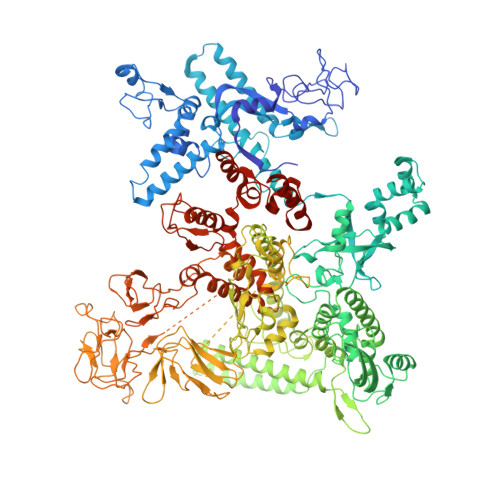
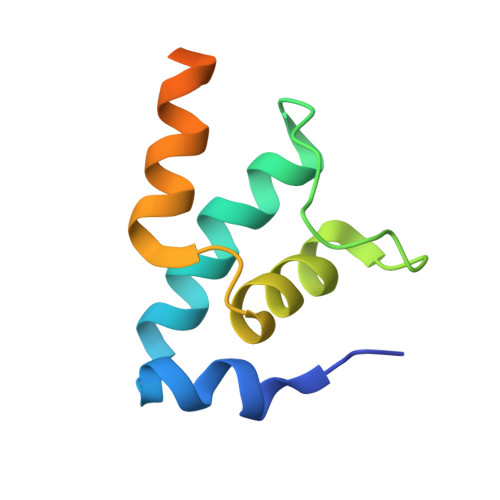
Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
He, D., You, L., Wu, X., Shi, J., Wen, A., Yan, Z., Mu, W., Fang, C., Feng, Y., Zhang, Y.(2022) Nat Commun 13: 4204-4204
- PubMed: 35859063
- DOI: https://doi.org/10.1038/s41467-022-31871-7
- Primary Citation of Related Structures:
7F0R, 7VF9, 7XL3, 7XL4 - PubMed Abstract:
Pseudomonas aeruginosa (Pae) SutA adapts bacteria to hypoxia and nutrition-limited environment during chronic infection by increasing transcription activity of an RNA polymerase (RNAP) holoenzyme comprising the stress-responsive σ factor σ S (RNAP-σ S ). SutA shows no homology to previously characterized RNAP-binding proteins. The structure and mode of action of SutA remain unclear. Here we determined cryo-EM structures of Pae RNAP-σ S holoenzyme, Pae RNAP-σ S holoenzyme complexed with SutA, and Pae RNAP-σ S transcription initiation complex comprising SutA. The structures show SutA pinches RNAP-β protrusion and facilitates promoter unwinding by wedging RNAP-β lobe open. Our results demonstrate that SutA clears an energetic barrier to facilitate promoter unwinding of RNAP-σ S holoenzyme.
- Key Laboratory of Synthetic Biology, CAS Center for Excellence in Molecular Plant Sciences, Shanghai Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai, China.
Organizational Affiliation: