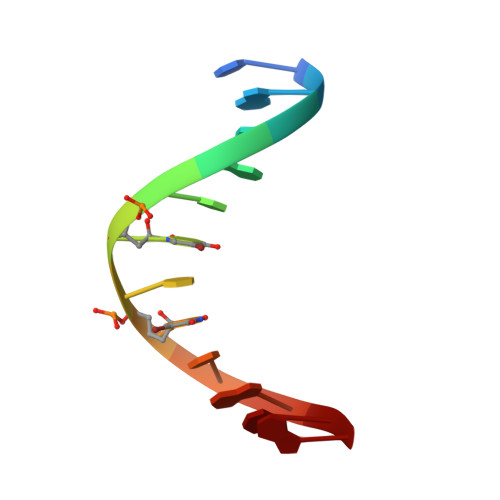
A Novel Ag I -DNA Rod Comprising a One-Dimensional Array of 11 Silver Ions within a Double Helical Structure.
Atsugi, T., Ono, A., Tasaka, M., Eguchi, N., Fujiwara, S., Kondo, J.(2022) Angew Chem Int Ed Engl 61: e202204798-e202204798
- PubMed: 35641885
- DOI: https://doi.org/10.1002/anie.202204798
- Primary Citation of Related Structures:
7XKM - PubMed Abstract:
DNA/RNA duplexes containing metal-ion-mediated base pairs (metallo-base pairs) have potential applications in developing nucleic acid-based nanodevices and genetic code expansion. Many metallo-base pairs are formed within duplexes stabilized by Watson-Crick base pairs. Recently, the crystal structure of an Ag I -DNA nanowire with an uninterrupted one-dimensional silver array was determined. Here, we present a new DNA helical wire, the "Ag I -DNA rod", containing an uninterrupted array of 11 Ag I ions. The Ag I -DNA rod consisted of only C-Ag I -C, G-Ag I -G, G-Ag I -5-bromouracil ( Br U), and Br U-Ag I - Br U metallo base pairs, with no Watson-Crick pairs. The Ag I -DNA rods were connected by non-canonical G-G pairs in crystals. Notably, data from our absorbance, circular dichroism, nuclear magnetic resonance, and mass spectrometry analyses suggested that the Ag I -DNA rods formed in solution, as well as within crystals.
- Department of Materials & Life Chemistry, Faculty of Engineering, Kanagawa University, 3-27-1 Rokkakubashi, Kanagawa-ku, Yokohama, 221-8686, Kanagawa, Japan.
Organizational Affiliation: