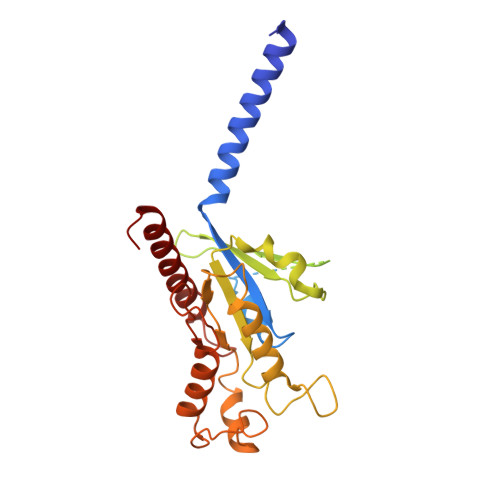
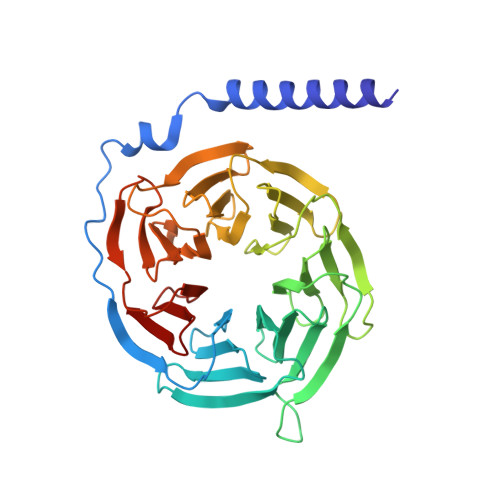
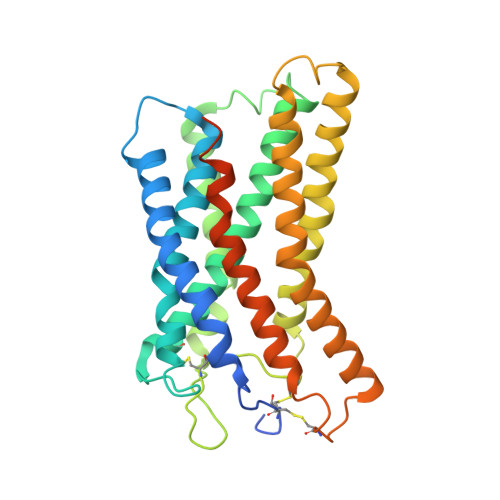
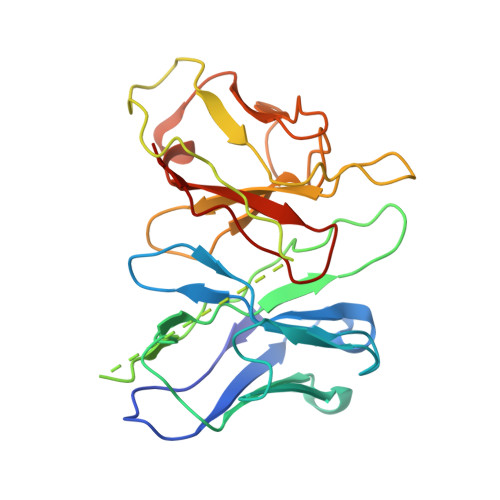
Structural insights into the human niacin receptor HCA2-G i signalling complex.
Yang, Y., Kang, H.J., Gao, R., Wang, J., Han, G.W., DiBerto, J.F., Wu, L., Tong, J., Qu, L., Wu, Y., Pileski, R., Li, X., Zhang, X.C., Zhao, S., Kenakin, T., Wang, Q., Stevens, R.C., Peng, W., Roth, B.L., Rao, Z., Liu, Z.J.(2023) Nat Commun 14: 1692-1692
- PubMed: 36973264
- DOI: https://doi.org/10.1038/s41467-023-37177-6
- Primary Citation of Related Structures:
7XK2, 7ZL9, 7ZLY - PubMed Abstract:
The hydroxycarboxylic acid receptor 2 (HCA2) agonist niacin has been used as treatment for dyslipidemia for several decades albeit with skin flushing as a common side-effect in treated individuals. Extensive efforts have been made to identify HCA2 targeting lipid lowering agents with fewer adverse effects, despite little being known about the molecular basis of HCA2 mediated signalling. Here, we report the cryo-electron microscopy structure of the HCA2-G i signalling complex with the potent agonist MK-6892, along with crystal structures of HCA2 in inactive state. These structures, together with comprehensive pharmacological analysis, reveal the ligand binding mode and activation and signalling mechanisms of HCA2. This study elucidates the structural determinants essential for HCA2 mediated signalling and provides insights into ligand discovery for HCA2 and related receptors.
- iHuman Institute, ShanghaiTech University, Shanghai, 201210, China.
Organizational Affiliation: