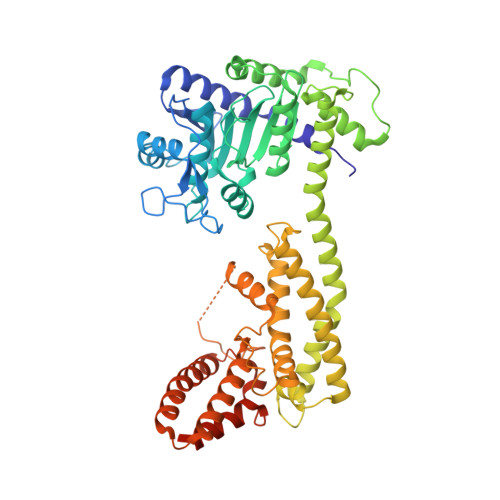
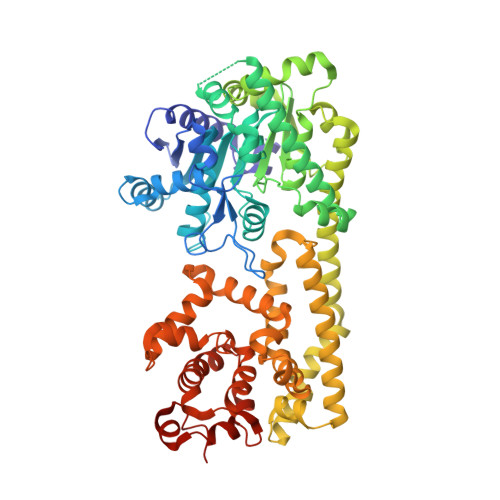
Identification and receptor mechanism of TIR-catalyzed small molecules in plant immunity.
Huang, S., Jia, A., Song, W., Hessler, G., Meng, Y., Sun, Y., Xu, L., Laessle, H., Jirschitzka, J., Ma, S., Xiao, Y., Yu, D., Hou, J., Liu, R., Sun, H., Liu, X., Han, Z., Chang, J., Parker, J.E., Chai, J.(2022) Science 377: eabq3297-eabq3297
- PubMed: 35857645
- DOI: https://doi.org/10.1126/science.abq3297
- Primary Citation of Related Structures:
7XDD, 7XEY - PubMed Abstract:
Plant nucleotide-binding leucine-rich repeat-containing (NLR) receptors with an N-terminal Toll/interleukin-1 receptor (TIR) domain sense pathogen effectors to enable TIR-encoded nicotinamide adenine dinucleotide hydrolase (NADase) activity for immune signaling. TIR-NLR signaling requires the helper NLRs N requirement gene 1 (NRG1), Activated Disease Resistance 1 (ADR1), and Enhanced Disease Susceptibility 1 (EDS1), which forms a heterodimer with each of its paralogs Phytoalexin Deficient 4 (PAD4) and Senescence-Associated Gene 101 (SAG101). Here, we show that TIR-containing proteins catalyze the production of 2'-(5''-phosphoribosyl)-5'-adenosine monophosphate (pRib-AMP) and diphosphate (pRib-ADP) in vitro and in planta. Biochemical and structural data demonstrate that EDS1-PAD4 is a receptor complex for pRib-AMP and pRib-ADP, which allosterically promote EDS1-PAD4 interaction with ADR1-L1 but not NRG1A. Our study identifies TIR-catalyzed pRib-AMP and pRib-ADP as a missing link in TIR signaling through EDS1-PAD4 and as likely second messengers for plant immunity.
- Beijing Advanced Innovation Center for Structural Biology, Tsinghua-Peking Joint Center for Life Sciences, Center for Plant Biology, School of Life Sciences, Tsinghua University, 100084 Beijing, China.
Organizational Affiliation: