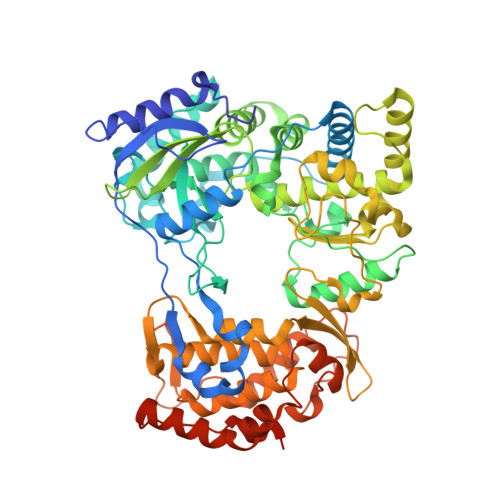
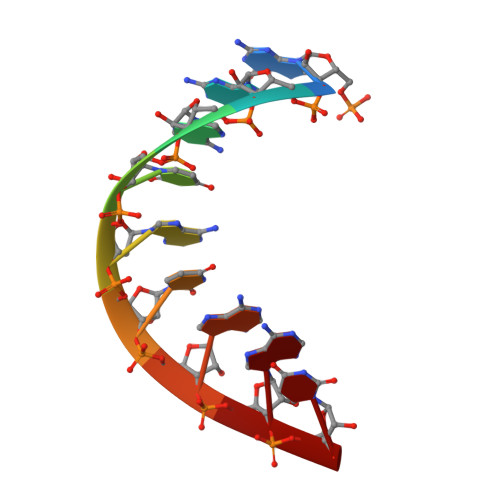
Structural basis of transition from initiation to elongation in de novo viral RNA-dependent RNA polymerases.
Wu, J., Wang, X., Liu, Q., Lu, G., Gong, P.(2023) Proc Natl Acad Sci U S A 120: e2211425120-e2211425120
- PubMed: 36577062
- DOI: https://doi.org/10.1073/pnas.2211425120
- Primary Citation of Related Structures:
7XD8, 7XD9 - PubMed Abstract:
De novo viral RNA-dependent RNA polymerases (RdRPs) utilize their priming element (PE) to facilitate accurate initiation. Upon transition to elongation, the PE has to retreat from the active site to give room to the template-product RNA duplex. However, PE conformational change upon this transition and the role of PE at elongation both remain elusive. Here, we report crystal structures of RdRP elongation complex (EC) from dengue virus serotype 2 (DENV2), demonstrating a dramatic refolding of PE that allows establishment of interactions with the RNA duplex backbone approved to be essential for EC stability. Enzymology data from both DENV2 and hepatitis C virus (HCV) RdRPs suggest that critical transition of the refolding likely occurs after synthesis of a 4- to 5-nucleotide (nt) product together providing a key basis in understanding viral RdRP transition from initiation to elongation.
- Key Laboratory of Special Pathogens and Biosafety, Center for Biosafety Mega-Science, Wuhan Institute of Virology, Chinese Academy of Sciences, Wuhan, Hubei 430071, China.
Organizational Affiliation: