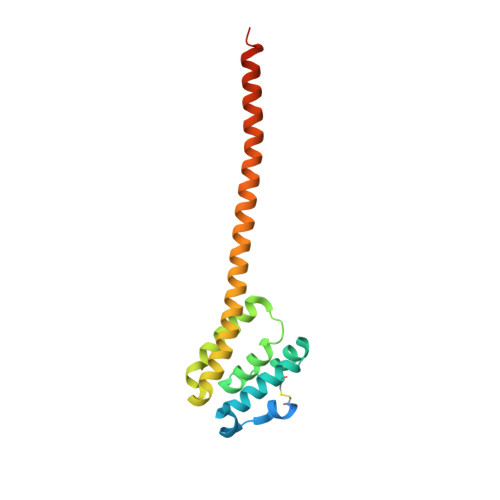
Crystal structures of QseE and QseG: elements of a three-component system from Escherichia coli.
Matsumoto, K., Fukuda, Y., Inoue, T.(2023) Acta Crystallogr F Struct Biol Commun 79: 285-293
- PubMed: 37877621
- DOI: https://doi.org/10.1107/S2053230X23009123
- Primary Citation of Related Structures:
7X6F, 7X6G, 7X6H, 8JWD - PubMed Abstract:
Bacteria regulate virulence by using two-component systems (TCSs) composed of a histidine kinase (HK) and a response regulator (RR). TCSs respond to environmental signals and change gene-expression levels. The HK QseE and the RR QseF regulate the virulence of Enterobacteriaceae bacteria such as enterohemorrhagic Escherichia coli. The operon encoding QseE/QseF also contains a gene encoding an outer membrane lipoprotein, qseG. The protein product QseG interacts with QseE in the periplasmic space to control the activity of QseE and constitutes a unique QseE/F/G three-component system. However, the structural bases of their functions are unknown. Here, crystal structures of the periplasmic regions of QseE and QseG were determined with the help of AlphaFold models. The periplasmic region of QseE has a helix-bundle structure as found in some HKs. The QseG structure is composed of an N-terminal globular domain and a long C-terminal helix forming a coiled-coil-like structure that contributes to dimerization. Comparison of QseG structures obtained from several crystallization conditions shows that QseG has structural polymorphisms at the C-terminus of the coiled-coil structure, indicating that the C-terminus is flexible. The C-terminal flexibility is derived from conserved hydrophilic residues that reduce the hydrophobic interaction at the coiled-coil interface. Electrostatic surface analysis suggests that the C-terminal coiled-coil region can interact with QseE. The observed structural fluctuation of the C-terminus of QseG is probably important for interaction with QseE.
- Graduate School of Pharmaceutical Science, Osaka University, Yamadaoka 1-6, Suita, Osaka 565-0871, Japan.
Organizational Affiliation: