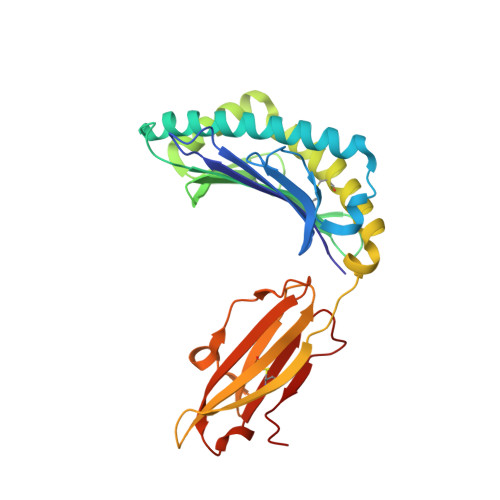
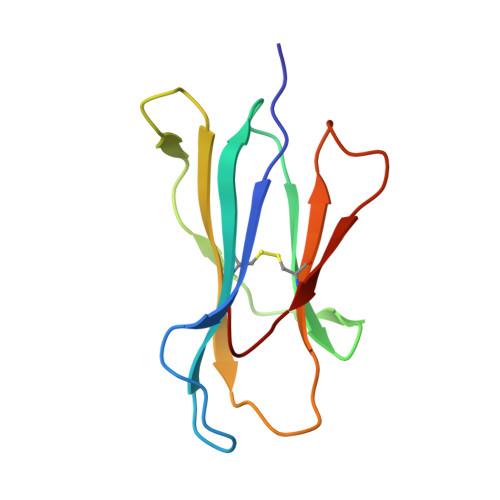
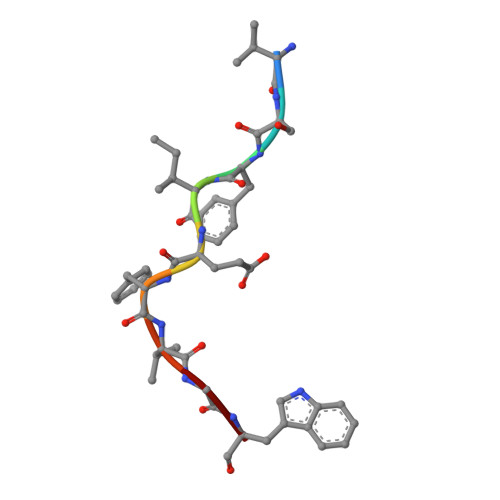
Allopurinol non-covalently facilitates binding of unconventional peptides to HLA-B*58:01.
Huan, X., Zhuo, N., Lee, H.Y., Ren, E.C.(2023) Sci Rep 13: 9373-9373
- PubMed: 37296297
- DOI: https://doi.org/10.1038/s41598-023-36293-z
- Primary Citation of Related Structures:
7WZZ, 7X00, 7X1B, 7X1C - PubMed Abstract:
Allopurinol, widely used in gout treatment, is the most common cause of severe cutaneous adverse drug reactions. The risk of developing such life-threatening reactions is increased particularly for HLA-B*58:01 positive individuals. However the mechanism of action between allopurinol and HLA remains unknown. We demonstrate here that a Lamin A/C peptide KAGQVVTI which is unable to bind HLA-B*58:01 on its own, is enabled to form a stable peptide-HLA complex only in the presence of allopurinol. Crystal structure analysis reveal that allopurinol non-covalently facilitated KAGQVVTI to adopt an unusual binding conformation, whereby the C-terminal isoleucine does not engage as a PΩ that typically fit deeply in the binding F-pocket. A similar observation, though to a lesser degree was seen with oxypurinol. Presentation of unconventional peptides by HLA-B*58:01 aided by allopurinol contributes to our fundamental understanding of drug-HLA interactions. The binding of peptides from endogenously available proteins such as self-protein lamin A/C and viral protein EBNA3B suggest that aberrant loading of unconventional peptides in the presence of allopurinol or oxypurinol may be able to trigger anti-self reactions that can lead to Stevens-Johnson syndrome/toxic epidermal necrolysis (SJS/TEN) and Drug Reaction with Eosinophilia and Systemic Symptoms (DRESS).
- Singapore Immunology Network (SigN), Agency for Science, Technology and Research (A*STAR), 8A Biomedical Grove, Immunos, Singapore, 138648, Singapore.
Organizational Affiliation: