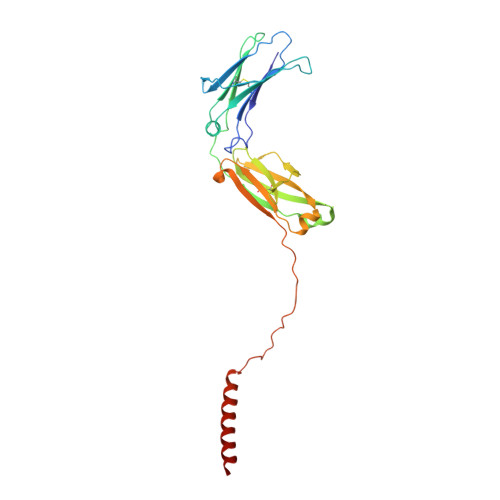
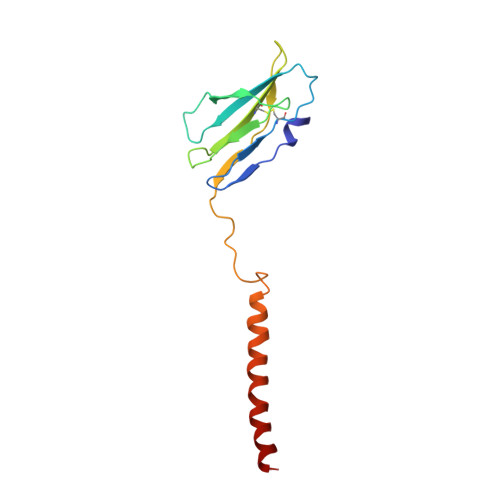
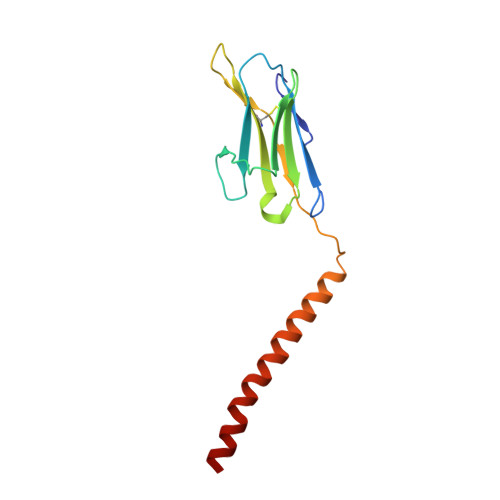
Cryo-EM structures of two human B cell receptor isotypes.
Ma, X., Zhu, Y., Chen, Y., Wang, S., Yang, D., Ma, Z., Zhang, A., Zhang, F., Guo, C., Huang, Z.(2022) Science 377: 880-885
- PubMed: 35981028
- DOI: https://doi.org/10.1126/science.abo3828
- Primary Citation of Related Structures:
7WSO, 7XT6 - PubMed Abstract:
The B cell receptor (BCR) complex plays a critical role in B cell development and immune responses. The assembly mechanisms underlying the BCR complex remain unknown. We determined the cryo-electron microscopy (cryo-EM) structures of human IgG-BCR and IgM-BCR, which consist of membrane-bound immunoglobulin molecules (mIg) and Igα/β subunits at a 1:1 stoichiometry. Assembly of both BCR complexes involves their extracellular domains, membrane-proximal connection peptides, and transmembrane (TM) helices. The TM helices of mIgG and mIgM share a conserved set of hydrophobic and polar interactions with Igα/β TM helices. By contrast, the IgG-Cγ3 and IgM-Cμ4 domains interact with extracellular Ig-like domains of Igα/β through head-to-tail and side-by-side modes, respectively. This work reveals the structural basis for BCR assembly and provides insights into BCR triggering.
- HIT Center for Life Sciences, School of Life Science and Technology, Harbin Institute of Technology, Harbin 150080, China.
Organizational Affiliation: