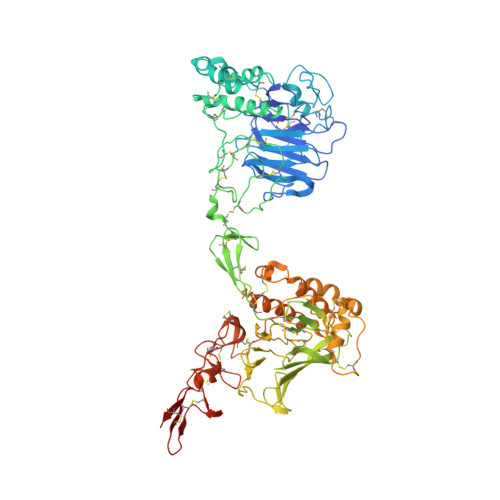
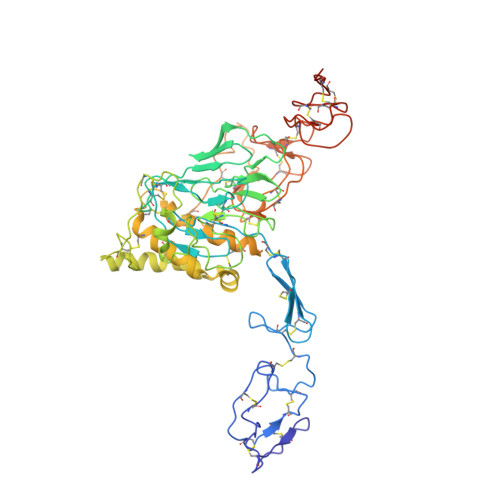
Structural mechanism of VWF D'D3 dimer formation.
Shu, Z., Zeng, J., Xia, L., Cai, H., Zhou, A.(2022) Cell Discov 8: 14-14
- PubMed: 35169124
- DOI: https://doi.org/10.1038/s41421-022-00378-2
- Primary Citation of Related Structures:
7WN3, 7WN4, 7WN6 - Department of Pathophysiology, Key Laboratory of Cell Differentiation and Apoptosis of Chinese Ministry of Education, Shanghai Jiao Tong University School of Medicine, Shanghai, China.
Organizational Affiliation: