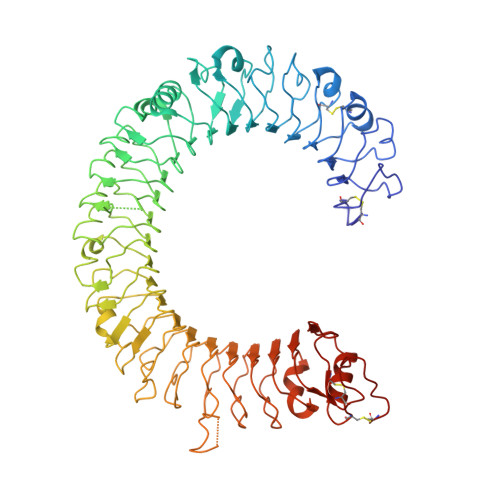
TLR3 forms a laterally aligned multimeric complex along double-stranded RNA for efficient signal transduction.
Sakaniwa, K., Fujimura, A., Shibata, T., Shigematsu, H., Ekimoto, T., Yamamoto, M., Ikeguchi, M., Miyake, K., Ohto, U., Shimizu, T.(2023) Nat Commun 14: 164-164
- PubMed: 36631495
- DOI: https://doi.org/10.1038/s41467-023-35844-2
- Primary Citation of Related Structures:
7WM4 - PubMed Abstract:
Toll-like receptor 3 (TLR3) is a member of the TLR family, which plays an important role in the innate immune system and is responsible for recognizing viral double-stranded RNA (dsRNA). Previous biochemical and structural studies have revealed that a minimum length of approximately 40-50 base pairs of dsRNA is necessary for TLR3 binding and dimerization. However, efficient TLR3 activation requires longer dsRNA and the molecular mechanism underlying its dsRNA length-dependent activation remains unknown. Here, we report cryo-electron microscopy analyses of TLR3 complexed with longer dsRNA. TLR3 dimers laterally form a higher multimeric complex along dsRNA, providing the basis for cooperative binding and efficient signal transduction.
- Graduate School of Pharmaceutical Sciences, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo, 113-0033, Japan.
Organizational Affiliation: