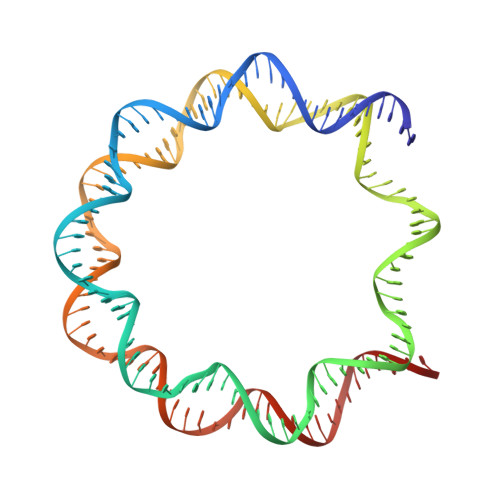
Structural and biochemical analyses of the nucleosome containing Komagataella pastoris histones.
Fukushima, Y., Hatazawa, S., Hirai, S., Kujirai, T., Ehara, H., Sekine, S.I., Takizawa, Y., Kurumizaka, H.(2022) J Biochem 172: 79-88
- PubMed: 35485963
- DOI: https://doi.org/10.1093/jb/mvac043
- Primary Citation of Related Structures:
7WLR - PubMed Abstract:
Komagataella pastoris is a methylotrophic yeast that is commonly used as a host cell for protein production. In the present study, we reconstituted the nucleosome with K. pastoris histones and determined the structure of the nucleosome core particle by cryogenic electron microscopy. In the K. pastoris nucleosome, the histones form an octamer and the DNA is left-handedly wrapped around it. Micrococcal nuclease assays revealed that the DNA ends of the K. pastoris nucleosome are somewhat more accessible, as compared with those of the human nucleosome. In vitro transcription assays demonstrated that the K. pastoris nucleosome is transcribed by the K. pastoris RNA polymerase II (RNAPII) more efficiently than the human nucleosome, while the RNAPII pausing positions of the K. pastoris nucleosome are the same as those of the human nucleosome. These results suggested that the DNA end flexibility may enhance the transcription efficiency in the nucleosome but minimally affect the nucleosomal pausing positions of RNAPII.
- Laboratory of Chromatin Structure and Function, Institute for Quantitative Biosciences, The University of Tokyo, 1-1-1 Yayoi, Bunkyo-ku, Tokyo 113-0032, Japan.
Organizational Affiliation: