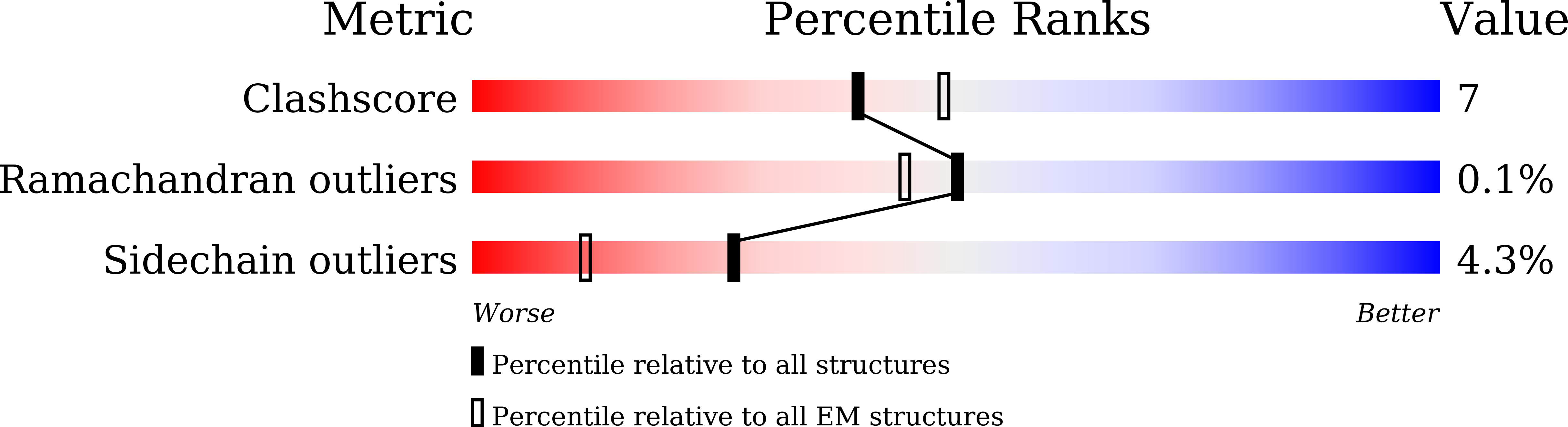Asymmetric pendrin homodimer reveals its molecular mechanism as anion exchanger.
Liu, Q., Zhang, X., Huang, H., Chen, Y., Wang, F., Hao, A., Zhan, W., Mao, Q., Hu, Y., Han, L., Sun, Y., Zhang, M., Liu, Z., Li, G.L., Zhang, W., Shu, Y., Sun, L., Chen, Z.(2023) Nat Commun 14: 3012-3012
- PubMed: 37230976
- DOI: https://doi.org/10.1038/s41467-023-38303-0
- Primary Citation of Related Structures:
7WK1, 7WK7, 7WL2, 7WL7, 7WL8, 7WL9, 7WLA, 7WLB, 7WLE - PubMed Abstract:
Pendrin (SLC26A4) is an anion exchanger expressed in the apical membranes of selected epithelia. Pendrin ablation causes Pendred syndrome, a genetic disorder associated with sensorineural hearing loss, hypothyroid goiter, and reduced blood pressure. However its molecular structure has remained unknown, limiting our understanding of the structural basis of transport. Here, we determine the cryo-electron microscopy structures of mouse pendrin with symmetric and asymmetric homodimer conformations. The asymmetric homodimer consists of one inward-facing protomer and the other outward-facing protomer, representing coincident uptake and secretion- a unique state of pendrin as an electroneutral exchanger. The multiple conformations presented here provide an inverted alternate-access mechanism for anion exchange. The structural and functional data presented here disclose the properties of an anion exchange cleft and help understand the importance of disease-associated variants, which will shed light on the pendrin exchange mechanism.
- The Fifth People's Hospital of Shanghai and Institutes of Biomedical Sciences, Fudan University, Shanghai, 200032, China.
Organizational Affiliation: