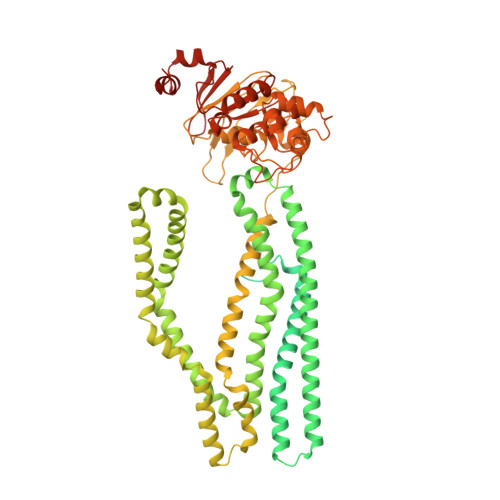
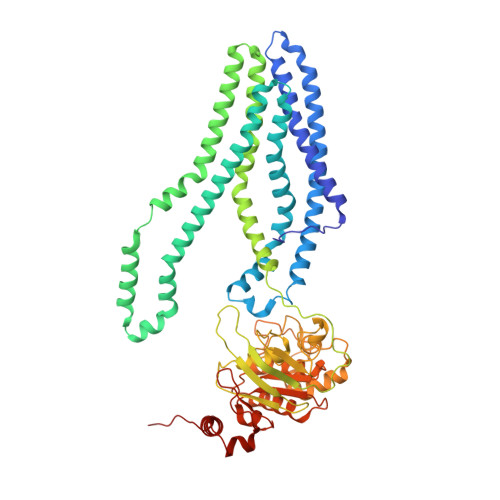
Cryo-EM structures for the Mycobacterium tuberculosis iron-loaded siderophore transporter IrtAB.
Sun, S., Gao, Y., Yang, X., Yang, X., Hu, T., Liang, J., Xiong, Z., Ran, Y., Ren, P., Bai, F., Guddat, L.W., Yang, H., Rao, Z., Zhang, B.(2023) Protein Cell 14: 448-458
- PubMed: 36882106
- DOI: https://doi.org/10.1093/procel/pwac060
- Primary Citation of Related Structures:
7WIU, 7WIV, 7WIW, 7WIX - PubMed Abstract:
The adenosine 5'-triphosphate (ATP)-binding cassette (ABC) transporter, IrtAB, plays a vital role in the replication and viability of Mycobacterium tuberculosis (Mtb), where its function is to import iron-loaded siderophores. Unusually, it adopts the canonical type IV exporter fold. Herein, we report the structure of unliganded Mtb IrtAB and its structure in complex with ATP, ADP, or ATP analogue (AMP-PNP) at resolutions ranging from 2.8 to 3.5 Å. The structure of IrtAB bound ATP-Mg2+ shows a "head-to-tail" dimer of nucleotide-binding domains (NBDs), a closed amphipathic cavity within the transmembrane domains (TMDs), and a metal ion liganded to three histidine residues of IrtA in the cavity. Cryo-electron microscopy (Cryo-EM) structures and ATP hydrolysis assays show that the NBD of IrtA has a higher affinity for nucleotides and increased ATPase activity compared with IrtB. Moreover, the metal ion located in the TM region of IrtA is critical for the stabilization of the conformation of IrtAB during the transport cycle. This study provides a structural basis to explain the ATP-driven conformational changes that occur in IrtAB.
- Shanghai Institute for Advanced Immunochemical Studies and School of Life Science and Technology, ShanghaiTech University, Shanghai 201210, China.
Organizational Affiliation: