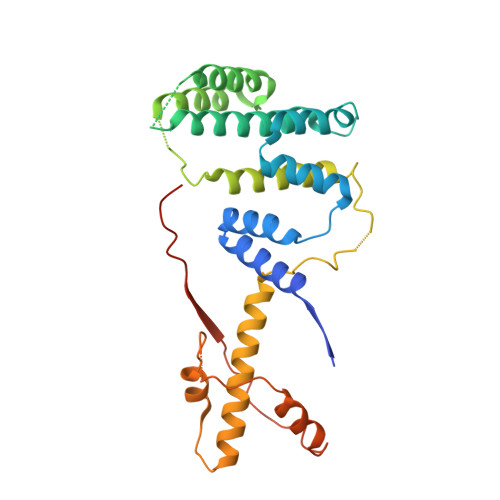
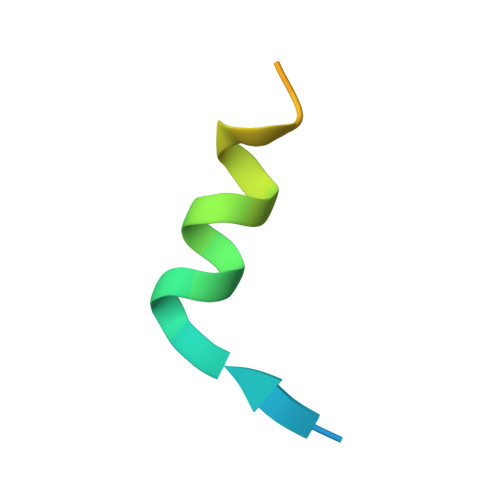
Intertwined Wdr47-NTD dimer recognizes a basic-helical motif in Camsap proteins for proper central-pair microtubule formation.
Ren, J., Li, D., Liu, J., Liu, H., Yan, X., Zhu, X., Feng, W.(2022) Cell Rep 41: 111589-111589
- PubMed: 36351391
- DOI: https://doi.org/10.1016/j.celrep.2022.111589
- Primary Citation of Related Structures:
7WEJ, 7WEK - PubMed Abstract:
Calmodulin-regulated spectrin-associated proteins (Camsaps) bind to the N-terminal domain of WD40-repeat 47 (Wdr47-NTD; featured with a LisH-CTLH motif) to properly generate axonemal central-pair microtubules (CP-MTs) for the planar beat pattern of mammalian motile multicilia. The underlying molecular mechanism, however, remains unclear. Here, we determine the structures of apo-Wdr47-NTD and Wdr47-NTD in complex with a characteristic Wdr47-binding region (WBR) from Camsap3. Wdr47-NTD forms an intertwined dimer with a special cross-over region (COR) in addition to the canonical LisH and globular α-helical core (GAC). The basic WBR peptide adopts an α-helical conformation and anchors to a tailored acidic pocket embedded in the COR. Mutations in this target-binding pocket disrupt the interaction between Wdr47-NTD and Camsap3. Impairing Wdr47-Camsap interactions markedly reduces rescue effects of Wdr47 on CP-MTs and ciliary beat of Wdr47-deficient ependymal cells. Thus, Wdr47-NTD functions by recognizing a specific basic helical motif in Camsap proteins via its non-canonical COR, a target-binding site in LisH-CTLH-containing domains.
- National Laboratory of Biomacromolecules, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, 15 Datun Road, Beijing 100101, China.
Organizational Affiliation: