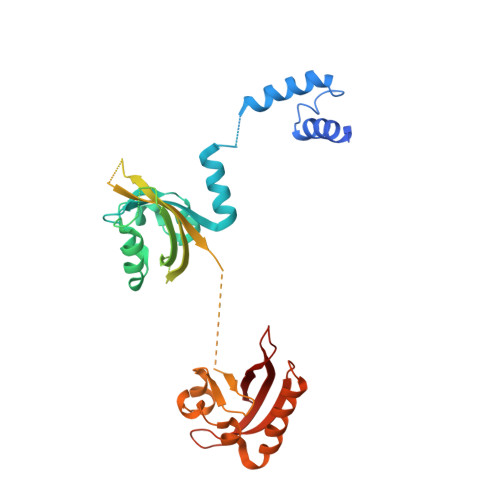
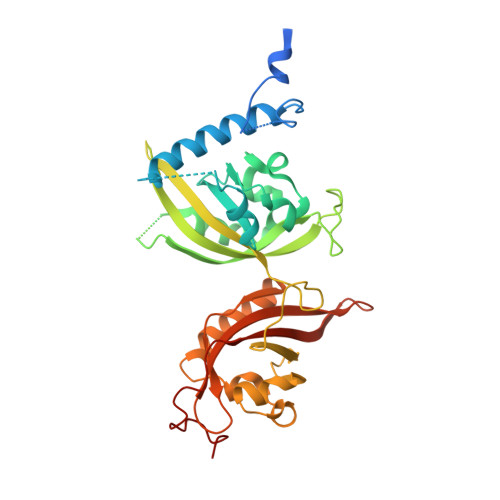
Structural basis for the allosteric inhibition of hypoxia-inducible factor (HIF)-2 by belzutifan.
Ren, X., Diao, X., Zhuang, J., Wu, D.(2022) Mol Pharmacol
- PubMed: 36167425
- DOI: https://doi.org/10.1124/molpharm.122.000525
- Primary Citation of Related Structures:
7W80 - PubMed Abstract:
Hypoxia-inducible factor (HIF)-2α and its obligate heterodimerization partner aryl hydrocarbon receptor nuclear translocator (ARNT), are both members of the basic helix-loop-helix-PER-ARNT-SIM (bHLH-PAS) transcription factor family. Previous studies have identified HIF-2α as a key oncogenic driver in clear cell renal cell carcinoma (ccRCC), rendering it a promising drug target for this type of kidney cancer. Belzutifan is the first HIF-2α inhibitor approved for treating ccRCC and other cancers associated with the von Hippel-Lindau (VHL) disease. However, the detailed inhibitory mechanism of belzutifan at molecular level is still unclear. Here we obtained the crystal structure of HIF-2α-ARNT heterodimer in complex with belzutifan at 2.75 Å resolution. The complex structure shows that belzutifan binds into the PAS-B pocket of HIF-2α, and it destabilizes the dimerization of HIF-2α and ARNT through allosteric effects mainly mediated by the key residue M252 of HIF-2α near the dimer interface. We further explored the inhibitory effects of belzutifan using biochemical and functional assays. The time-resolved fluorescence energy transfer (TR-FRET)-based binding assay showed that belzutifan disrupts the dimerization of HIF-2α and ARNT with a K i value of 20 nM. The luciferase reporter assay indicated that belzutifan can efficiently inhibit the transcriptional activity of HIF-2α with an IC 50 value of 17 nM. Besides, the real-time PCR assay illustrated that belzutifan can reduce the expression of HIF-2α downstream genes in 786-O kidney cancer cells in a dose-dependent manner. Our work reveals the molecular mechanism by which belzutifan allosterically inhibits HIF-2α and provides valuable information for the subsequent drug development targeting HIF-2α. Significance Statement The bHLH-PAS family of transcription factors are an emerging group of small-molecule drug targets. Belzutifan, originally developed by Peloton Therapeutics, is the first FDA-approved drug directly binding to a bHLH-PAS protein, the hypoxia-inducible factor (HIF)-2α. Based on the protein-drug complex structure, biochemical binding assays, and functional profiling of downstream gene expression, this study reveals the regulatory mechanism of how belzutifan allosterically destabilizes HIF-2α's heterodimerization with its obligate partner protein, thus reducing their transcriptional activity that links to tumor progression.
- State Key Laboratory of Microbial Technology, Shandong University, China.
Organizational Affiliation: