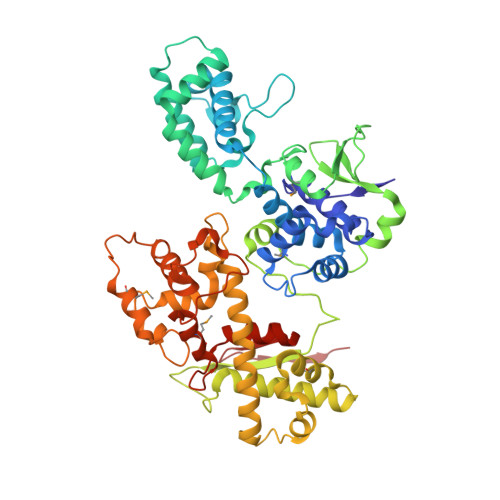
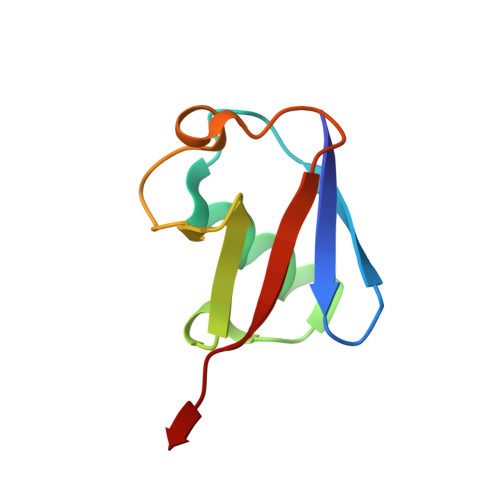
Structural basis for the dual catalytic activity of the Legionella pneumophila ovarian tumor (OTU) domain deubiquitinase LotA.
Luo, J., Ruan, X., Huang, Z., Li, Z., Ye, L., Wu, Y., Zhen, X., Ouyang, S.(2022) J Biological Chem 298: 102414-102414
- PubMed: 36007613
- DOI: https://doi.org/10.1016/j.jbc.2022.102414
- Primary Citation of Related Structures:
7W54 - PubMed Abstract:
Legionella pneumophila, a bacterial pathogen that causes a severe pneumonia known as Legionnaires' disease, extensively exploits the ubiquitin (Ub) pathway in the infected host cells through certain virulence effectors excreted by the Dot/Icm system. To date, several Dot/Icm effectors have been found to act as Ub ligases, and four effectors, including LotA, LotB, LotC, and Ceg7, have been identified as deubiquitinases (DUBs) from the ovarian tumor (OTU) domain family. LotA is unique among other OTU DUBs because it possesses two distinct DUB domains and exclusively exhibits catalytic activity against K6-linked diUb and polyUb chains. However, the structure of LotA and the molecular mechanism for the dual DUB activity remains elusive. In this study, we solved the structure of LotA in complex with proximally bound Ub and distal covalently bound Ub. Both Ub molecules are bound to the DUB1 domain and mimic a K6-linked diUb. Structural analysis reveals that the DUB1 domain utilizes a distinct mechanism for recognition of the K6-linked diUb within a large S1' binding site that is uncommon to OTU DUBs. Structural fold of the LotA DUB2 domain closely resembles LotB and LotC, similarly containing an extra α-helix lobe that has been demonstrated to play an important role in Ub binding. Collectively, our study uncovers the structural basis for the dual catalytic activity of the unique OTU family DUB LotA.
- Provincial University Key Laboratory of Cellular Stress Response and Metabolic Regulation, The Key Laboratory of Innate Immune Biology of Fujian Province, Biomedical Research Center of South China, Key Laboratory of OptoElectronic Science and Technology for Medicine of the Ministry of Education, College of Life Sciences, Fujian Normal University, Fuzhou, China.
Organizational Affiliation: