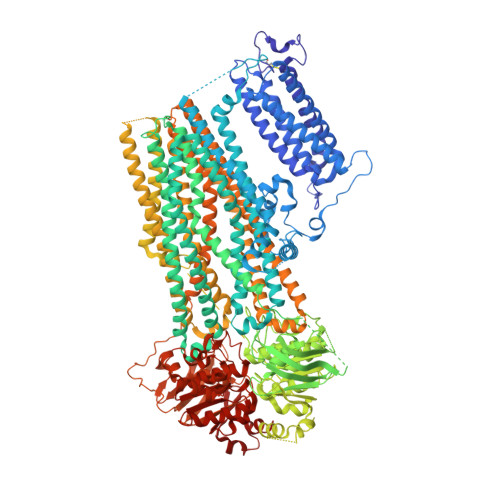
Structural insights into the mechanism of pancreatic K ATP channel regulation by nucleotides.
Wang, M., Wu, J.X., Ding, D., Chen, L.(2022) Nat Commun 13: 2770-2770
- PubMed: 35589716
- DOI: https://doi.org/10.1038/s41467-022-30430-4
- Primary Citation of Related Structures:
7W4O, 7W4P - PubMed Abstract:
ATP-sensitive potassium channels (K ATP ) are metabolic sensors that convert the intracellular ATP/ADP ratio to the excitability of cells. They are involved in many physiological processes and implicated in several human diseases. Here we present the cryo-EM structures of the pancreatic K ATP channel in both the closed state and the pre-open state, resolved in the same sample. We observe the binding of nucleotides at the inhibitory sites of the Kir6.2 channel in the closed but not in the pre-open state. Structural comparisons reveal the mechanism for ATP inhibition and Mg-ADP activation, two fundamental properties of K ATP channels. Moreover, the structures also uncover the activation mechanism of diazoxide-type K ATP openers.
- State Key Laboratory of Membrane Biology, College of Future Technology, Institute of Molecular Medicine, Peking University, Beijing Key Laboratory of Cardiometabolic Molecular Medicine, 100871, Beijing, China.
Organizational Affiliation: