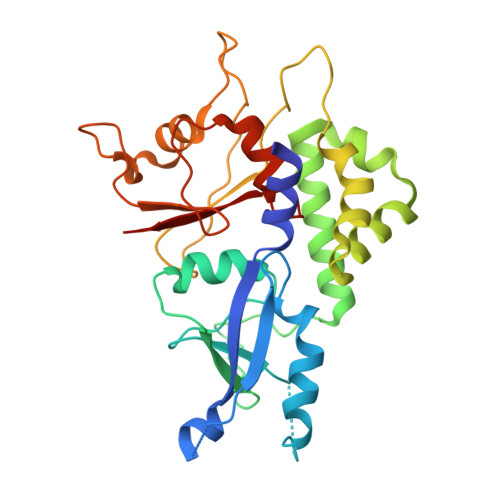
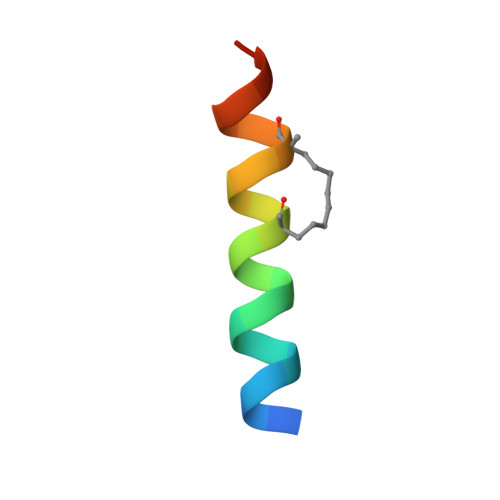
Targeting the ATG5-ATG16L1 Protein-Protein Interaction with a Hydrocarbon-Stapled Peptide Derived from ATG16L1 for Autophagy Inhibition.
Cui, J., Ogasawara, Y., Kurata, I., Matoba, K., Fujioka, Y., Noda, N.N., Shibasaki, M., Watanabe, T.(2022) J Am Chem Soc 144: 17671-17679
- PubMed: 36107218
- DOI: https://doi.org/10.1021/jacs.2c07648
- Primary Citation of Related Structures:
7W36 - PubMed Abstract:
Selective modulation of autophagy is a promising therapeutic strategy, especially for cancer treatment. However, the lack of specific autophagy inhibitors limits this strategy. The formation of the ATG12-ATG5-ATG16L1 complex is essential for targeting the ATG12-ATG5 conjugate to proper membranes and to generate LC3-II for the progression of autophagy. Thus, targeting ATG5-ATG16L1 protein-protein interactions (PPIs) might inhibit early stage autophagy with high specificity. In this paper, we report that a stapled peptide derived from ATG16L1 exhibits potent binding affinity to ATG5, striking resistance to proteolysis, and significant autophagy inhibition activities in cells.
- Institute of Microbial Chemistry (BIKAKEN), Tokyo, 3-14-23 Kamiosaki Shinagawa-ku, Tokyo, 141-0021, Japan.
Organizational Affiliation: