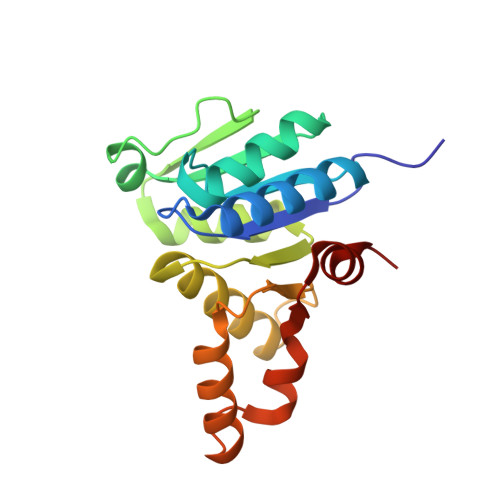
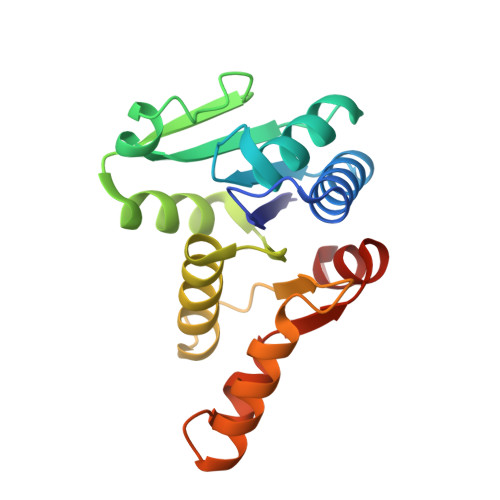
Crystal structure of the cytokinin-producing enzyme "lonely guy" (LOG) from Mycobacterium tuberculosis.
Shang, L., Li, G., Lin, Q., Ou, M., Liang, J., Xiao, G., Wang, Z., Cui, S., Zhang, T., Liu, L., Zhang, G.(2022) Biochem Biophys Res Commun 598: 113-118
- PubMed: 35158209
- DOI: https://doi.org/10.1016/j.bbrc.2022.01.103
- Primary Citation of Related Structures:
7W2I - PubMed Abstract:
Mycobacterium tuberculosis (Mtb) is an extremely successful intracellular pathogen that cause a large number of death worldwide. It is interesting that this non-phytopathogen can synthesize cytokinin by "lonely guy" (LOG) protein. The cytokinin biosynthesis pathway in Mtb is not clear. Here we determined the crystal structure of LOG from Mtb (MtLOG) at a high resolution of 1.8 Å. MtLOG exists as dimer which belongs to type-I LOG and shows a typical α-β Rossmann fold. Like other LOGs, MtLOG also contains a conserved "PGGXGTXXE" motif that contributes to the formation of an active site. For the first time, we found that the MtLOG binds to Mg 2+ in the negative potential pocket. According to the docking result, we found that Arg78, Arg98 and Tyr162 should be the key amino acid involved in substrate binding. Our findings provide a structural basis for cytokinin study in Mtb and will play an important role in design and development of enzyme inhibitors.
- National Clinical Research Center for Infectious Diseases, Guangdong Provincial Clinical Research Center for Tuberculosis, Shenzhen Third People's Hospital, Southern University of Science and Technology, Shenzhen, 518112, China.
Organizational Affiliation: