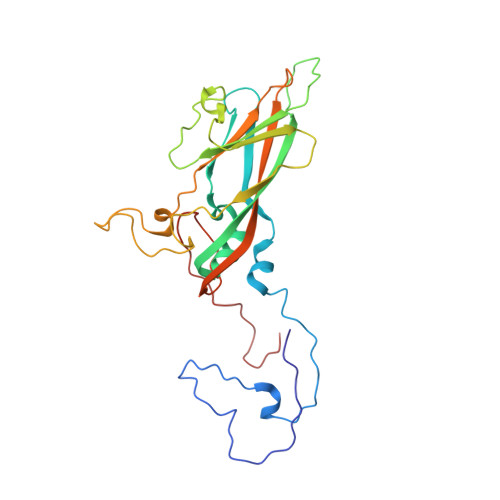
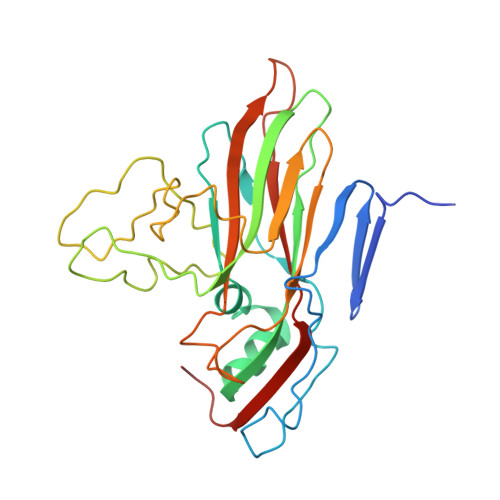
Molecular basis of differential receptor usage for naturally occurring CD55-binding and -nonbinding coxsackievirus B3 strains.
Wang, Q., Yang, Q., Liu, C., Wang, G., Song, H., Shang, G., Peng, R., Qu, X., Liu, S., Cui, Y., Wang, P., Xu, W., Zhao, X., Qi, J., Yang, M., Gao, G.F.(2022) Proc Natl Acad Sci U S A 119
- PubMed: 35046043
- DOI: https://doi.org/10.1073/pnas.2118590119
- Primary Citation of Related Structures:
7VXH, 7VXZ, 7VY0, 7VY5, 7VY6, 7VYK, 7VYL, 7VYM, 7W14, 7W17 - PubMed Abstract:
Receptor usage defines cell tropism and contributes to cell entry and infection. Coxsackievirus B (CVB) engages coxsackievirus and adenovirus receptor (CAR), and selectively utilizes the decay-accelerating factor (DAF; CD55) to infect cells. However, the differential receptor usage mechanism for CVB remains elusive. This study identified VP3-234 residues (234Q/N/V/D/E) as critical population selection determinants during CVB3 virus evolution, contributing to diverse binding affinities to CD55. Cryoelectron microscopy (cryo-EM) structures of CD55-binding/nonbinding isolates and their complexes with CD55 or CAR were obtained under both neutral and acidic conditions, and the molecular mechanism of VP3-234 residues determining CD55 affinity/specificity for naturally occurring CVB3 strains was elucidated. Structural and biochemical studies in vitro revealed the dynamic entry process of CVB3 and the function of the uncoating receptor CAR with different pH preferences. This work provides detailed insight into the molecular mechanism of CVB infection and contributes to an in-depth understanding of enterovirus attachment receptor usage.
- Department of Biomedical Sciences, City University of Hong Kong, Hong Kong SAR, China.
Organizational Affiliation: