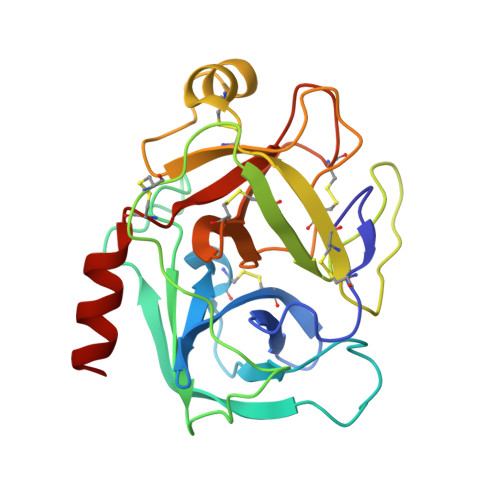
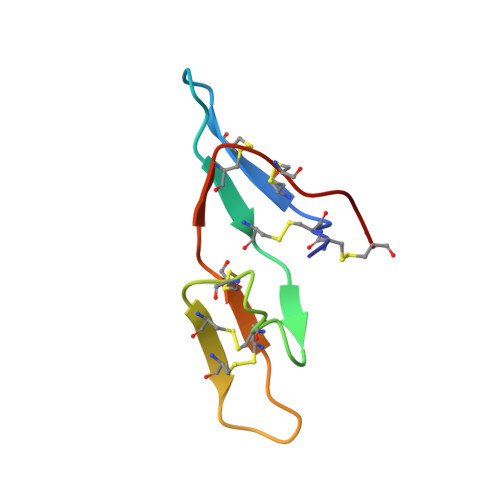
Insight into the structural basis of the dual inhibitory mode of Lima bean (Phaseolus lunatus) serine protease inhibitor.
Ahmad, M.S., Akbar, Z., Choudhary, M.I.(2023) Proteins 91: 22-31
- PubMed: 35927030
- DOI: https://doi.org/10.1002/prot.26407
- Primary Citation of Related Structures:
7VO7 - PubMed Abstract:
Bovine pancreatic trypsin was crystallized, in-complex with Lima bean trypsin inhibitor (LBTI) (Phaseolus lunatus L.), in the form of a ternary complex. LBTI is a Bowman-Birk-type bifunctional serine protease inhibitor, which has two independent inhibitory loops. Both of the loops can inhibit trypsin, however, only the hydrophobic loop is specific for inhibiting chymotrypsin. The structure of trypsin incomplex with the LBTI has been solved and refined at 2.25 Å resolution, in the space group P4 1, with R work /R free values of 18.1/23.3. The two binding sites of LBTI differ in only two amino acids. Lysine and leucine are the key residues of the two different binding loops positioned at the P1, and involved in binding the S1 binding site of trypsin. The asymmetric unit cell contains two molecules of trypsin and one molecule of LBTI. The key interactions include hydrogen bonds between LBTI and active site residues of trypsin. The 3D structure of the enzyme-inhibitor complex provided details insight into the trypsin inhibition by LBTI. To the best of our knowledge, this is the first report on the structure of trypsin incomplex with LBTI.
- Dr. Panjwani Center for Molecular Medicine and Drug Research, International Center for Chemical and Biological Sciences, University of Karachi, Karachi, Pakistan.
Organizational Affiliation: