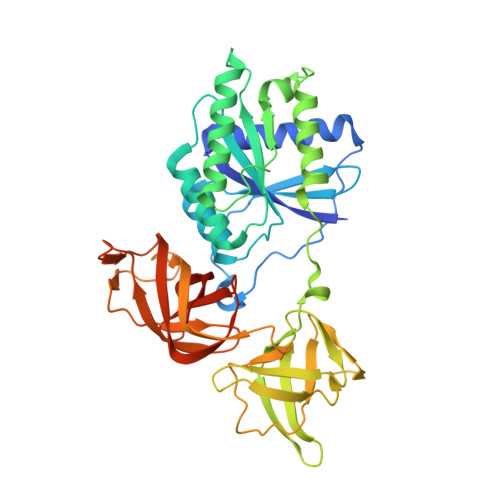
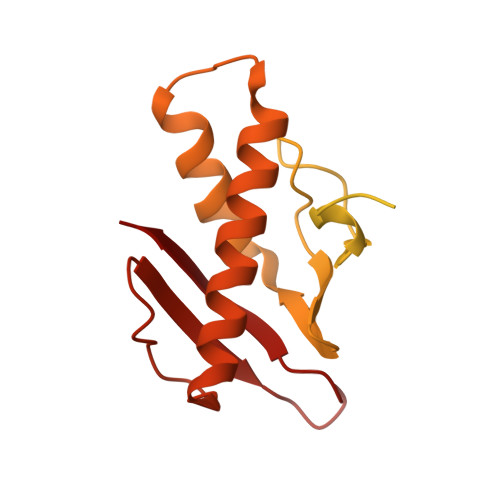
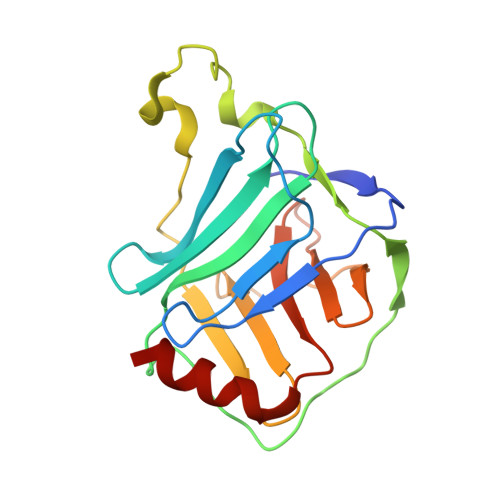
Mechanistic insights into tRNA cleavage by a contact-dependent growth inhibitor protein and translation factors.
Wang, J., Yashiro, Y., Sakaguchi, Y., Suzuki, T., Tomita, K.(2022) Nucleic Acids Res 50: 4713-4731
- PubMed: 35411396
- DOI: https://doi.org/10.1093/nar/gkac228
- Primary Citation of Related Structures:
7VMC - PubMed Abstract:
Contact-dependent growth inhibition is a mechanism of interbacterial competition mediated by delivery of the C-terminal toxin domain of CdiA protein (CdiA-CT) into neighboring bacteria. The CdiA-CT of enterohemorrhagic Escherichia coli EC869 (CdiA-CTEC869) cleaves the 3'-acceptor regions of specific tRNAs in a reaction that requires the translation factors Tu/Ts and GTP. Here, we show that CdiA-CTEC869 has an intrinsic ability to recognize a specific sequence in substrate tRNAs, and Tu:Ts complex promotes tRNA cleavage by CdiA-CTEC869. Uncharged and aminoacylated tRNAs (aa-tRNAs) were cleaved by CdiA-CTEC869 to the same extent in the presence of Tu/Ts, and the CdiA-CTEC869:Tu:Ts:tRNA(aa-tRNA) complex formed in the presence of GTP. CdiA-CTEC869 interacts with domain II of Tu, thereby preventing the 3'-moiety of tRNA to bind to Tu as in canonical Tu:GTP:aa-tRNA complexes. Superimposition of the Tu:GTP:aa-tRNA structure onto the CdiA-CTEC869:Tu structure suggests that the 3'-portion of tRNA relocates into the CdiA-CTEC869 active site, located on the opposite side to the CdiA-CTEC869 :Tu interface, for tRNA cleavage. Thus, CdiA-CTEC869 is recruited to Tu:GTP:Ts, and CdiA-CT:Tu:GTP:Ts recognizes substrate tRNAs and cleaves them. Tu:GTP:Ts serves as a reaction scaffold that increases the affinity of CdiA-CTEC869 for substrate tRNAs and induces a structural change of tRNAs for efficient cleavage by CdiA-CTEC869.
- Department of Computational Biology and Medical Sciences, Graduate School of Frontier Sciences, The University of Tokyo, Kashiwa,Chiba277-8562, Japan.
Organizational Affiliation: