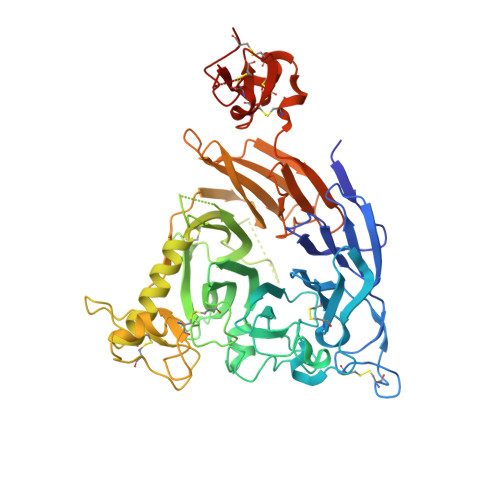
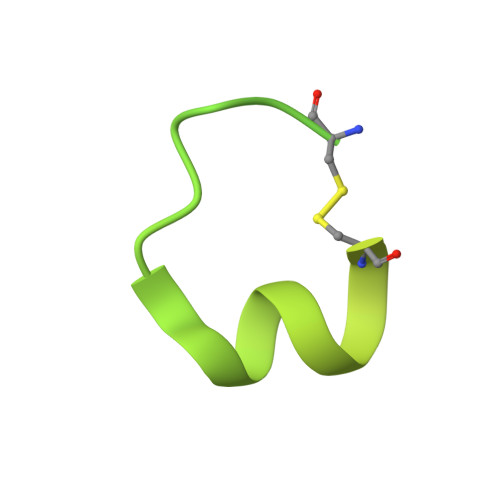
De novo Fc-based receptor dimerizers differentially modulate PlexinB1 function.
Sugano-Nakamura, N., Matoba, K., Hirose, M., Bashiruddin, N.K., Matsunaga, Y., Yamashita, K., Hirata, K., Yamamoto, M., Arimori, T., Suga, H., Takagi, J.(2022) Structure 30: 1411-1423.e4
- PubMed: 35981535
- DOI: https://doi.org/10.1016/j.str.2022.07.008
- Primary Citation of Related Structures:
7VF3, 7VG7 - PubMed Abstract:
Signaling by single-pass transmembrane receptors often involves a formation of ligand-induced receptor dimers with particular conformation, and bivalent receptor binders can modulate receptor functions by inducing different receptor dimer conformations, although such agents are difficult to design. Here, we describe the generation of both antagonistic and agonistic receptor dimerizers toward PlexinB1 (PlxnB1), a receptor for semaphorin 4D (Sema4D), by grafting two different PlxnB1-binding peptides onto the human immunoglobulin G1 (IgG1) Fc protein. The function-modulating activity of a peptide Fc was strongly dependent on the type of the peptide as well as the grafting site, with the best variants showing activity at an nM concentration range. Structural analysis of each peptide-PlxnB1 complex revealed that the agonistic Fc dimerizes PlxnB1 in a face-to-face fashion similar to that induced by Sema4D, whereas antagonistic Fc would induce signaling-incompetent PlxnB1 dimer conformation, enforcing the idea that plexin activation is primarily controlled by the receptor orientation within the dimer.
- Laboratory for Protein Synthesis and Expression, Institute for Protein Research, Osaka University, Osaka, Japan.
Organizational Affiliation: