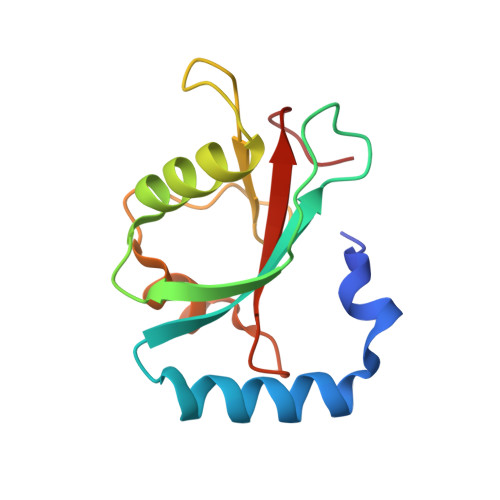
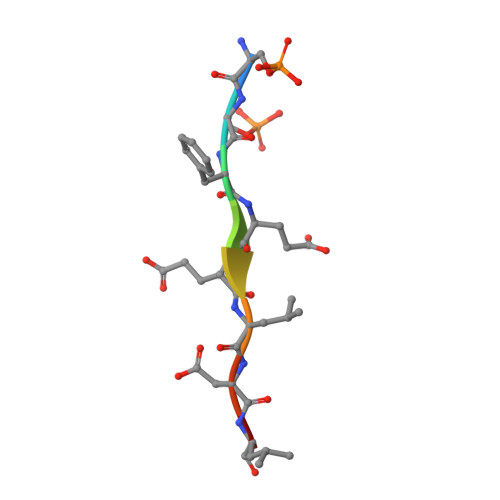
Phosphorylation by casein kinase 2 enhances the interaction between ER-phagy receptor TEX264 and ATG8 proteins.
Chino, H., Yamasaki, A., Ode, K.L., Ueda, H.R., Noda, N.N., Mizushima, N.(2022) EMBO Rep 23: e54801-e54801
- PubMed: 35417087
- DOI: https://doi.org/10.15252/embr.202254801
- Primary Citation of Related Structures:
7VEC, 7VED - PubMed Abstract:
Selective autophagy cargos are recruited to autophagosomes primarily by interacting with autophagosomal ATG8 family proteins via the LC3-interacting region (LIR). The upstream sequence of most LIRs contains negatively charged residues such as Asp, Glu, and phosphorylated Ser and Thr. However, the significance of LIR phosphorylation (compared with having acidic amino acids) and the structural basis of phosphorylated LIR-ATG8 binding are not entirely understood. Here, we show that the serine residues upstream of the core LIR of the endoplasmic reticulum (ER)-phagy receptor TEX264 are phosphorylated by casein kinase 2, which is critical for its interaction with ATG8s, autophagosomal localization, and ER-phagy. Structural analysis shows that phosphorylation of these serine residues increases binding affinity by producing multiple hydrogen bonds with ATG8s that cannot be mimicked by acidic residues. This binding mode is different from those of other ER-phagy receptors that utilize a downstream helix, which is absent from TEX264, to increase affinity. These results suggest that phosphorylation of the LIR is critically important for strong LIR-ATG8 interactions, even in the absence of auxiliary interactions.
- Department of Biochemistry and Molecular Biology, Graduate School of Medicine, The University of Tokyo, Bunkyo-ku, Tokyo, Japan.
Organizational Affiliation: