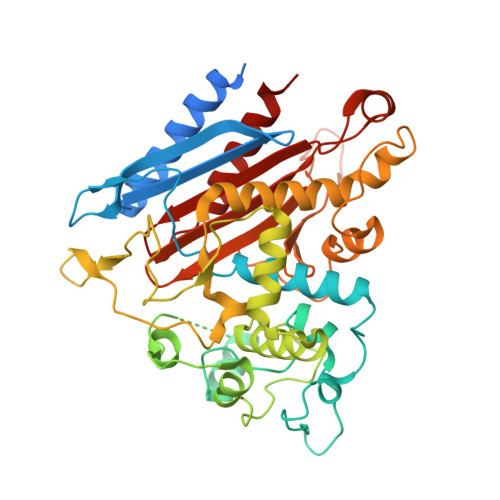
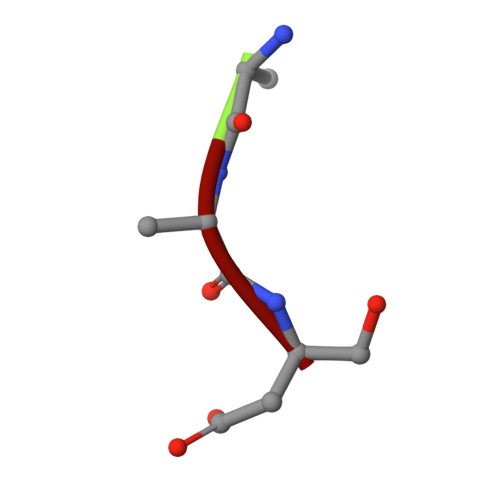
Structural basis for the catalytic activity of filamentous human serine beta-lactamase-like protein LACTB.
Zhang, M., Zhang, L., Guo, R., Xiao, C., Yin, J., Zhang, S., Yang, M.(2022) Structure 30: 685-696.e5
- PubMed: 35247327
- DOI: https://doi.org/10.1016/j.str.2022.02.007
- Primary Citation of Related Structures:
7V1Y, 7V1Z, 7V21 - PubMed Abstract:
Serine beta-lactamase-like protein (LACTB) is a mammalian mitochondrial serine protease that can specifically hydrolyze peptide bonds adjacent to aspartic acid residues and is structurally related to prokaryotic penicillin-binding proteins. Here, we determined the cryoelectron microscopy structures of human LACTB (hLACTB) filaments from wild-type protein, a middle region deletion mutant, and in complex with the inhibitor Z-AAD-CMK at 3.0-, 3.1-, and 2.8-Å resolution, respectively. Structural analysis and activity assays revealed that three interfaces are required for the assembly of hLACTB filaments and that the formation of higher order helical structures facilitates its cleavage activity. Further structural and enzymatic analyses of middle region deletion constructs indicated that, while this region is necessary for substrate hydrolysis, it is not required for filament formation. Moreover, the inhibitor-bound structure showed that hLACTB may cleave peptide bonds adjacent to aspartic acid residues. These findings provide the structural basis underlying hLACTB catalytic activity.
- Ministry of Education Key Laboratory of Protein Science, Tsinghua-Peking Center for Life Sciences, Beijing Advanced Innovation Center for Structural Biology, School of Life Sciences, Tsinghua University, Beijing 100084, China.
Organizational Affiliation: