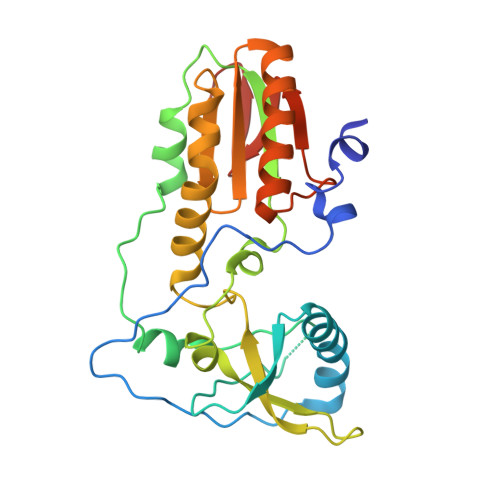
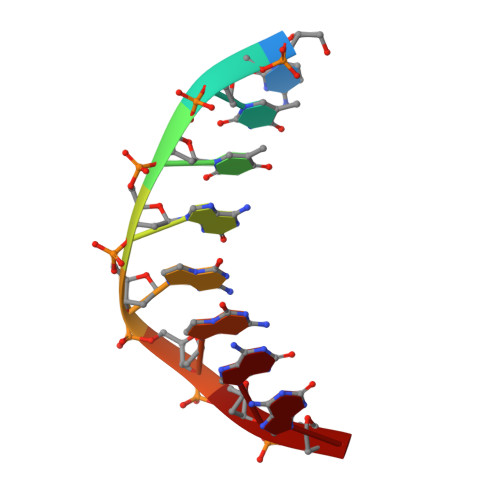
Crystal structures of 'ALternative Isoinformational ENgineered' DNA in B-form.
Shukla, M.S., Hoshika, S., Benner, S.A., Georgiadis, M.M.(2023) Philos Trans R Soc Lond B Biol Sci 378: 20220028-20220028
- PubMed: 36633282
- DOI: https://doi.org/10.1098/rstb.2022.0028
- Primary Citation of Related Structures:
7UYN, 7UYO, 7UYP - PubMed Abstract:
The first structural model of duplex DNA reported in 1953 by Watson & Crick presented the double helix in B-form, the form that genomic DNA exists in much of the time. Thus, artificial DNA seeking to mimic the properties of natural DNA should also be able to adopt B-form. Using a host-guest system in which Moloney murine leukemia virus reverse transcriptase serves as the host and DNA as the guests, we determined high-resolution crystal structures of three complexes including 5'-CTT BPPBBSSZZS AAG, 5'-CTT SSPBZPSZBB AAG and 5'-CTT ZZPBSBSZPP AAG with 10 consecutive unnatural nucleobase pairs in B-form within self-complementary 16 bp duplex oligonucleotides. We refer to this ALternative Isoinformational ENgineered (ALIEN) genetic system containing two nucleobase pairs ( P:Z , pairing 2-amino-imidazo-[1,2- a ]-1,3,5-triazin-(8 H )-4-one with 6-amino-5-nitro-(1 H )-pyridin-2-one, and B:S , 6-amino-4-hydroxy-5-(1 H )-purin-2-one with 3-methyl-6-amino-pyrimidin-2-one) as ALIEN DNA. We characterized both position- and sequence-specific helical, nucleobase pair and dinucleotide step parameters of P:Z and B:S pairs in the context of B-form DNA. We conclude that ALIEN DNA exhibits structural features that vary with sequence. Further, Z can participate in alternative stacking modes within a similar sequence context as captured in two different structures. This finding suggests that ALIEN DNA may have a larger repertoire of B-form structures than natural DNA. This article is part of the theme issue 'Reactivity and mechanism in chemical and synthetic biology'.
- Department of Biochemistry and Molecular Biology, Indiana University School of Medicine, Indianapolis, IN 46202, USA.
Organizational Affiliation: