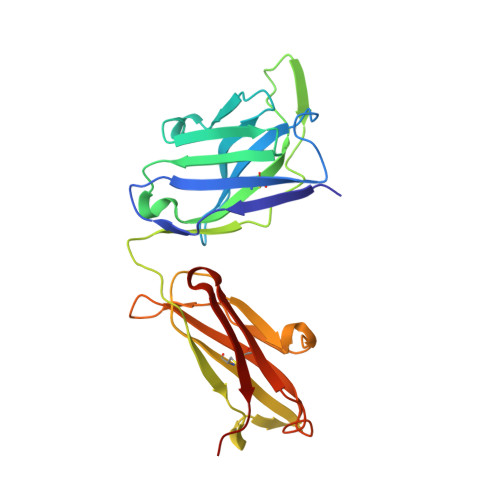
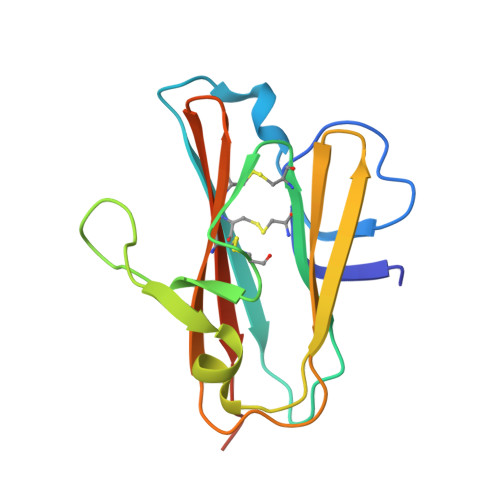
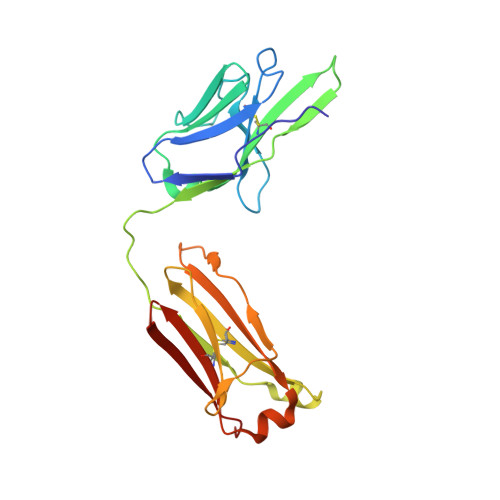
Highly potent, naturally acquired human monoclonal antibodies against Pfs48/45 block Plasmodium falciparum transmission to mosquitoes.
Fabra-Garcia, A., Hailemariam, S., de Jong, R.M., Janssen, K., Teelen, K., van de Vegte-Bolmer, M., van Gemert, G.J., Ivanochko, D., Semesi, A., McLeod, B., Vos, M.W., de Bruijni, M.H.C., Bolscher, J.M., Szabat, M., Vogt, S., Kraft, L., Duncan, S., Kamya, M.R., Feeney, M.E., Jagannathan, P., Greenhouse, B., Dechering, K.J., Sauerwein, R.W., King, C.R., MacGill, R.S., Bousema, T., Julien, J.P., Jore, M.M.(2023) Immunity 56: 406-419.e7
- PubMed: 36792574
- DOI: https://doi.org/10.1016/j.immuni.2023.01.009
- Primary Citation of Related Structures:
7UXL - PubMed Abstract:
Malaria transmission-blocking vaccines (TBVs) aim to induce antibodies that interrupt malaria parasite development in the mosquito, thereby blocking onward transmission, and provide a much-needed tool for malaria control and elimination. The parasite surface protein Pfs48/45 is a leading TBV candidate. Here, we isolated and characterized a panel of 81 human Pfs48/45-specific monoclonal antibodies (mAbs) from donors naturally exposed to Plasmodium parasites. Genetically diverse mAbs against each of the three domains (D1-D3) of Pfs48/45 were identified. The most potent mAbs targeted D1 and D3 and achieved >80% transmission-reducing activity in standard membrane-feeding assays, at 10 and 2 μg/mL, respectively. Co-crystal structures of D3 in complex with four different mAbs delineated two conserved protective epitopes. Altogether, these Pfs48/45-specific human mAbs provide important insight into protective and non-protective epitopes that can further our understanding of transmission and inform the design of refined malaria transmission-blocking vaccine candidates.
- Department of Medical Microbiology, Radboudumc, Nijmegen, the Netherlands.
Organizational Affiliation: