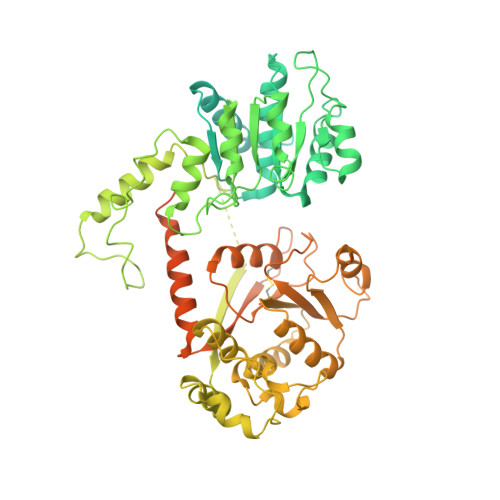
Chromatin remodeling of histone H3 variants by DDM1 underlies epigenetic inheritance of DNA methylation.
Lee, S.C., Adams, D.W., Ipsaro, J.J., Cahn, J., Lynn, J., Kim, H.S., Berube, B., Major, V., Calarco, J.P., LeBlanc, C., Bhattacharjee, S., Ramu, U., Grimanelli, D., Jacob, Y., Voigt, P., Joshua-Tor, L., Martienssen, R.A.(2023) Cell 186: 4100
- PubMed: 37643610
- DOI: https://doi.org/10.1016/j.cell.2023.08.001
- Primary Citation of Related Structures:
7UX9 - PubMed Abstract:
Nucleosomes block access to DNA methyltransferase, unless they are remodeled by DECREASE in DNA METHYLATION 1 (DDM1 LSH / HELLS), a Snf2-like master regulator of epigenetic inheritance. We show that DDM1 promotes replacement of histone variant H3.3 by H3.1. In ddm1 mutants, DNA methylation is partly restored by loss of the H3.3 chaperone HIRA, while the H3.1 chaperone CAF-1 becomes essential. The single-particle cryo-EM structure at 3.2 Å of DDM1 with a variant nucleosome reveals engagement with histone H3.3 near residues required for assembly and with the unmodified H4 tail. An N-terminal autoinhibitory domain inhibits activity, while a disulfide bond in the helicase domain supports activity. DDM1 co-localizes with H3.1 and H3.3 during the cell cycle, and with the DNA methyltransferase MET1 Dnmt1 , but is blocked by H4K16 acetylation. The male germline H3.3 variant MGH3/HTR10 is resistant to remodeling by DDM1 and acts as a placeholder nucleosome in sperm cells for epigenetic inheritance.
- Howard Hughes Medical Institute, Cold Spring Harbor Laboratory, 1 Bungtown Road, Cold Spring Harbor, NY 11724, USA.
Organizational Affiliation: