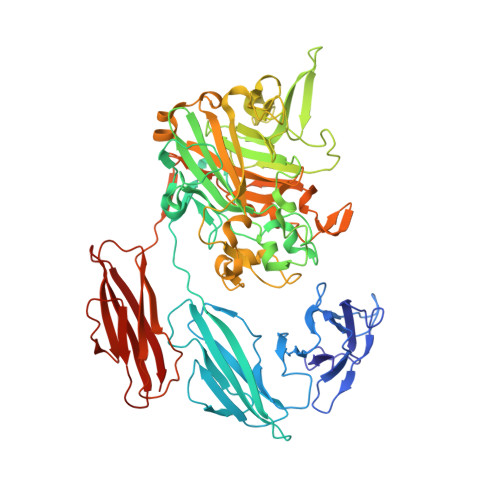
The Ruminococcus bromii amylosome protein Sas6 binds single and double helical alpha-glucan structures in starch.
Photenhauer, A.L., Villafuerte-Vega, R.C., Cerqueira, F.M., Armbruster, K.M., Marecek, F., Chen, T., Wawrzak, Z., Hopkins, J.B., Vander Kooi, C.W., Janecek, S., Ruotolo, B.T., Koropatkin, N.M.(2024) Nat Struct Mol Biol 31: 255-265
- PubMed: 38177679
- DOI: https://doi.org/10.1038/s41594-023-01166-6
- Primary Citation of Related Structures:
7UWU, 7UWV, 7UWW - PubMed Abstract:
Resistant starch is a prebiotic accessed by gut bacteria with specialized amylases and starch-binding proteins. The human gut symbiont Ruminococcus bromii expresses Sas6 (Starch Adherence System member 6), which consists of two starch-specific carbohydrate-binding modules from family 26 (RbCBM26) and family 74 (RbCBM74). Here, we present the crystal structures of Sas6 and of RbCBM74 bound with a double helical dimer of maltodecaose. The RbCBM74 starch-binding groove complements the double helical α-glucan geometry of amylopectin, suggesting that this module selects this feature in starch granules. Isothermal titration calorimetry and native mass spectrometry demonstrate that RbCBM74 recognizes longer single and double helical α-glucans, while RbCBM26 binds short maltooligosaccharides. Bioinformatic analysis supports the conservation of the amylopectin-targeting platform in CBM74s from resistant-starch degrading bacteria. Our results suggest that RbCBM74 and RbCBM26 within Sas6 recognize discrete aspects of the starch granule, providing molecular insight into how this structure is accommodated by gut bacteria.
- Department of Microbiology & Immunology, University of Michigan Medical School, Ann Arbor, MI, USA.
Organizational Affiliation: