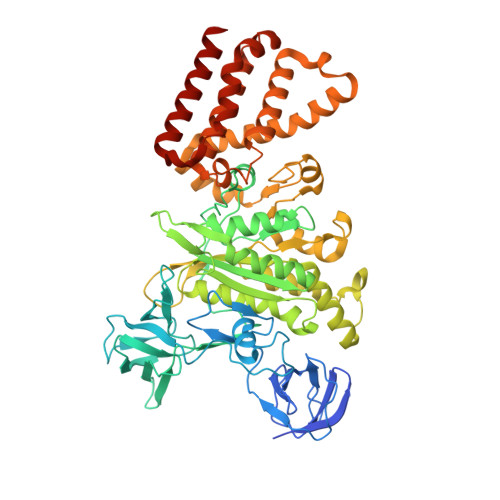
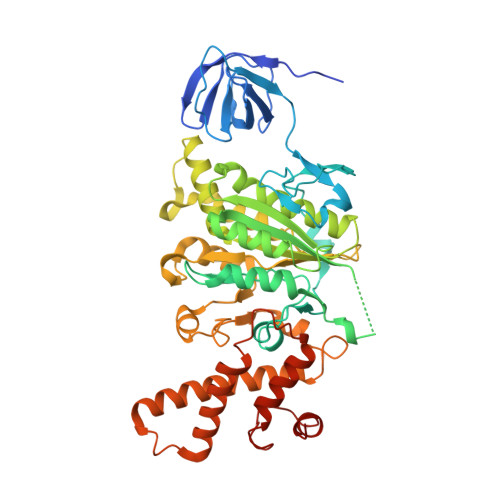
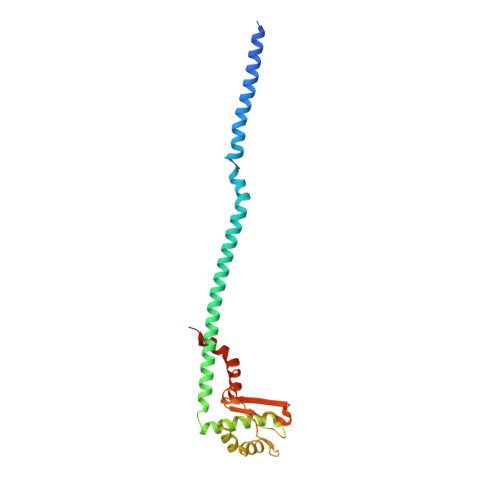
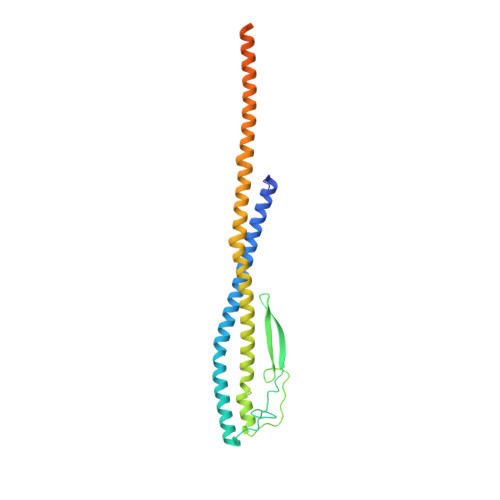
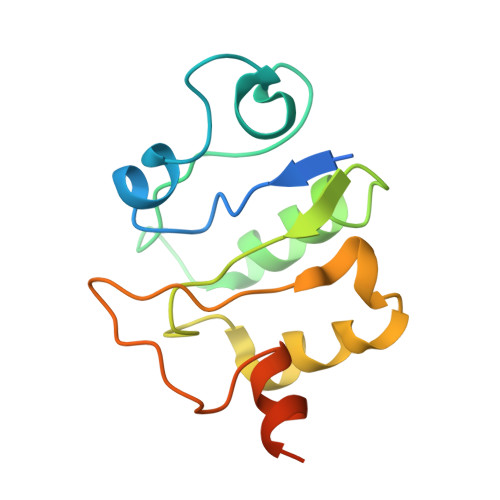
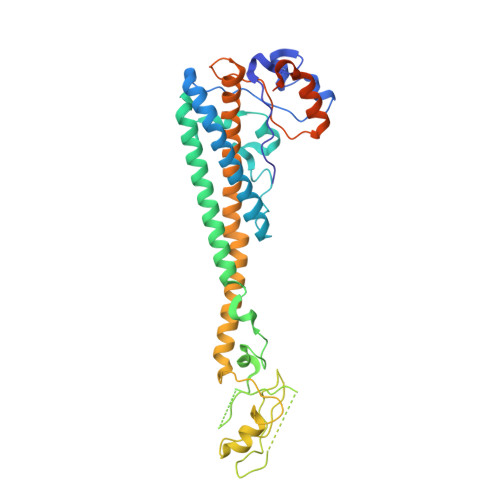
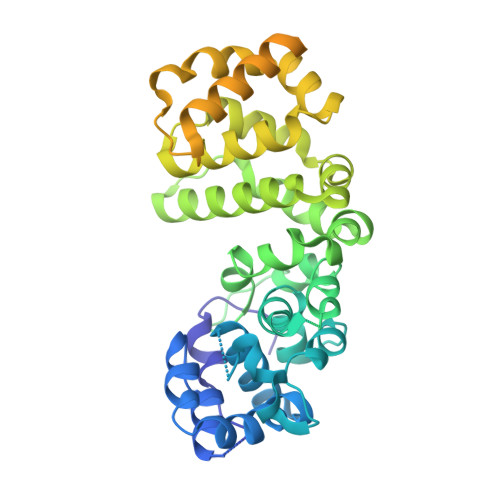
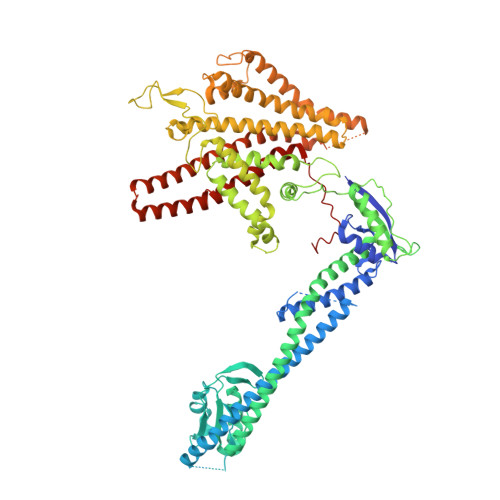
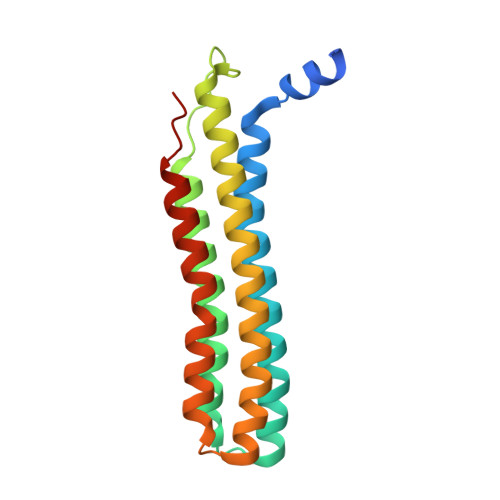
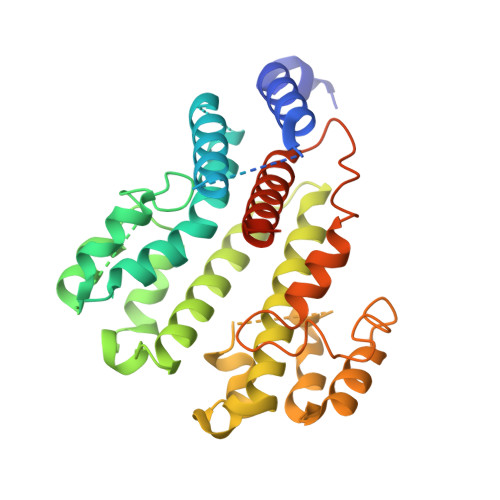
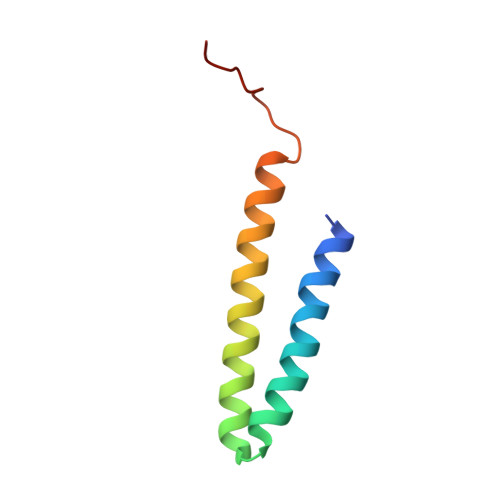
Structure of V-ATPase from citrus fruit.
Tan, Y.Z., Keon, K.A., Abdelaziz, R., Imming, P., Schulze, W., Schumacher, K., Rubinstein, J.L.(2022) Structure 30: 1403
- PubMed: 36041457
- DOI: https://doi.org/10.1016/j.str.2022.07.006
- Primary Citation of Related Structures:
7UW9, 7UWA, 7UWB, 7UWC, 7UWD - PubMed Abstract:
We used the Legionella pneumophila effector SidK to affinity purify the endogenous vacuolar-type ATPases (V-ATPases) from lemon fruit. The preparation was sufficient for cryoelectron microscopy, allowing structure determination of the enzyme in two rotational states. The structure defines the ATP:H + ratio of the enzyme, demonstrating that it can establish a maximum ΔpH of ∼3, which is insufficient to maintain the low pH observed in the vacuoles of juice sac cells in lemons and other citrus fruit. Compared with yeast and mammalian enzymes, the membrane region of the plant V-ATPase lacks subunit f and possesses an unusual configuration of transmembrane α helices. Subunit H, which inhibits ATP hydrolysis in the isolated catalytic region of V-ATPase, adopts two different conformations in the intact complex, hinting at a role in modulating activity in the intact enzyme.
- Molecular Medicine Program, The Hospital for Sick Children Research Institute, Toronto, ON M5G 0A4, Canada.
Organizational Affiliation: