S -adenosylhomocysteine analogs selectively suppress pan-coronavirus replication by inhibition of nsp14 methyltransferase.
Rosas-Lemus, M., Athe, S., Minasov, G., Pattie, J.A., Brunzelle, J.S., Chau, I., Li, F., Vedadi, M., Ma, H., Ramanathan, A., Becker, M.E., Hope, T.J., Abdelkarim, H., Grudzien, P., Gaponenko, V., Montgomery, J.E., Moellering, R.E., Rawal, V.H., Satchell, K.J.F.(2025) ACS Med Chem Lett
- PubMed: 41568342
- DOI: https://doi.org/10.1021/acsmedchemlett.5c00510
- Primary Citation of Related Structures:
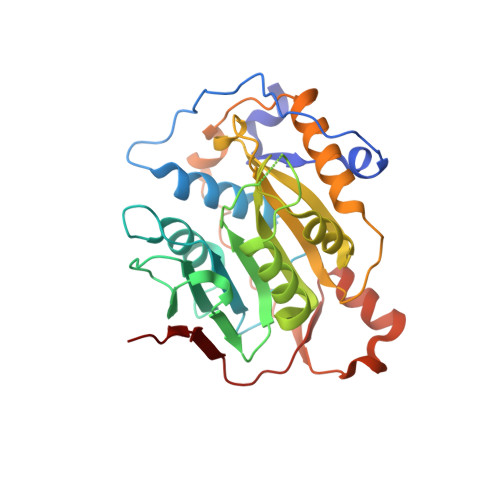
7ULT - PubMed Abstract:
To address the ongoing threat of SARS-CoV-2 and potential emergence of novel coronaviruses, we employed a comprehensive strategy to identify and synthesize inhibitors of coronavirus methyltransferases with chemical analogs of S -adenosylhomocysteine. Two analogs, designated 4h and 4p , inhibit both mouse hepatitis virus and SARS-CoV-2 replication. Compound 4p was most potent with half-maximal inhibition of biochemical activity at 0.2 μM and antiviral activity at ~20 μM. This compound also has low cytotoxicity and preferentially inhibits nsp14 over nsp16 and human methyltransferases. Furthermore, molecular docking based on a newly determined crystal structure of the apo nsp16-nsp10 complex predicts 4p occupies both the S -adenosylmethione and Gppp binding pockets of nsp14 and nsp16. Selectivity of 4p for nsp14 is likely due to enhanced structural stability of the nsp14 binding pocket relative to nsp16. These findings highlight SAH analogs as scaffolds for pan-coronavirus therapeutics and underscore the value of structure-guided design in antiviral drug discovery.
- Department of Microbiology-Immunology, Northwestern University Feinberg School of Medicine, Chicago, IL 60611 USA.
Organizational Affiliation: