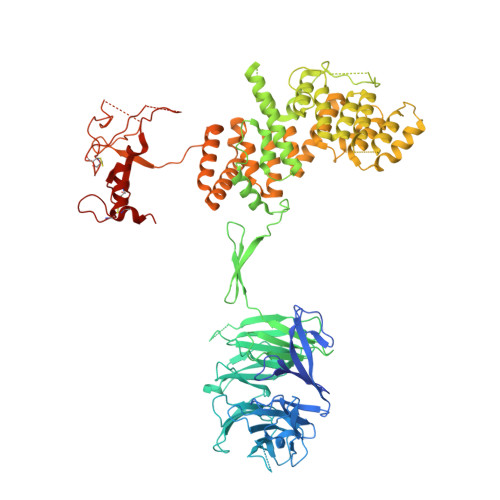
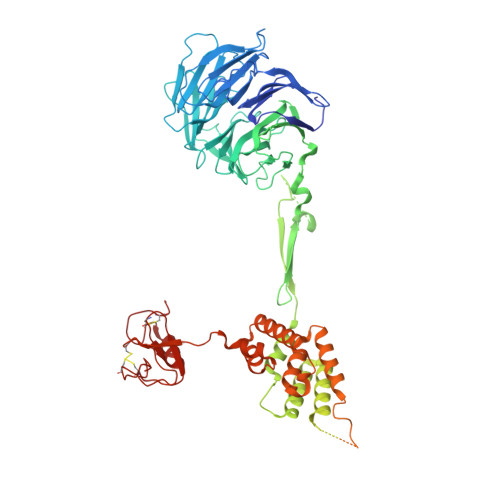
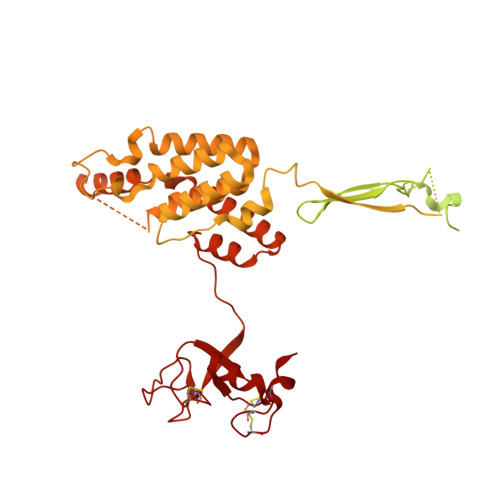
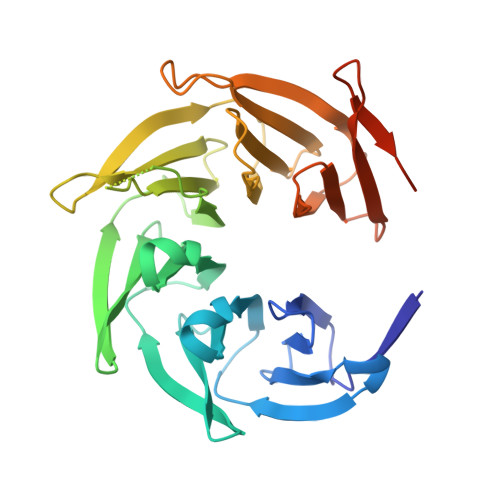
Structure of the nutrient-sensing hub GATOR2.
Valenstein, M.L., Rogala, K.B., Lalgudi, P.V., Brignole, E.J., Gu, X., Saxton, R.A., Chantranupong, L., Kolibius, J., Quast, J.P., Sabatini, D.M.(2022) Nature 607: 610-616
- PubMed: 35831510
- DOI: https://doi.org/10.1038/s41586-022-04939-z
- Primary Citation of Related Structures:
7UHY - PubMed Abstract:
Mechanistic target of rapamycin complex 1 (mTORC1) controls growth by regulating anabolic and catabolic processes in response to environmental cues, including nutrients 1,2 . Amino acids signal to mTORC1 through the Rag GTPases, which are regulated by several protein complexes, including GATOR1 and GATOR2. GATOR2, which has five components (WDR24, MIOS, WDR59, SEH1L and SEC13), is required for amino acids to activate mTORC1 and interacts with the leucine and arginine sensors SESN2 and CASTOR1, respectively 3-5 . Despite this central role in nutrient sensing, GATOR2 remains mysterious as its subunit stoichiometry, biochemical function and structure are unknown. Here we used cryo-electron microscopy to determine the three-dimensional structure of the human GATOR2 complex. We found that GATOR2 adopts a large (1.1 MDa), two-fold symmetric, cage-like architecture, supported by an octagonal scaffold and decorated with eight pairs of WD40 β-propellers. The scaffold contains two WDR24, four MIOS and two WDR59 subunits circularized via two distinct types of junction involving non-catalytic RING domains and α-solenoids. Integration of SEH1L and SEC13 into the scaffold through β-propeller blade donation stabilizes the GATOR2 complex and reveals an evolutionary relationship to the nuclear pore and membrane-coating complexes 6 . The scaffold orients the WD40 β-propeller dimers, which mediate interactions with SESN2, CASTOR1 and GATOR1. Our work reveals the structure of an essential component of the nutrient-sensing machinery and provides a foundation for understanding the function of GATOR2 within the mTORC1 pathway.
- Whitehead Institute for Biomedical Research, Cambridge, MA, USA. mvalenst@wi.mit.edu.
Organizational Affiliation: