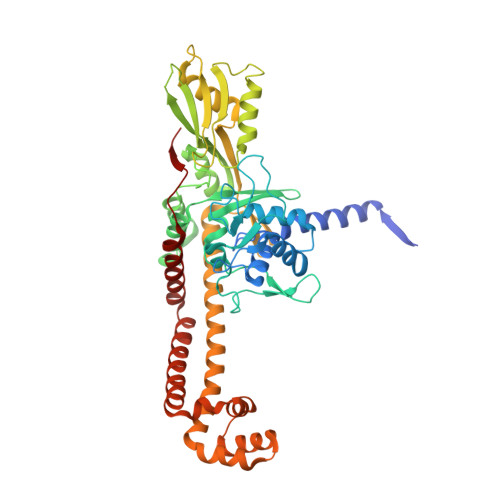
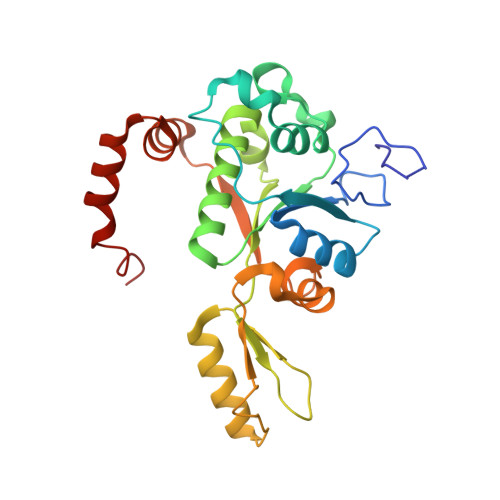
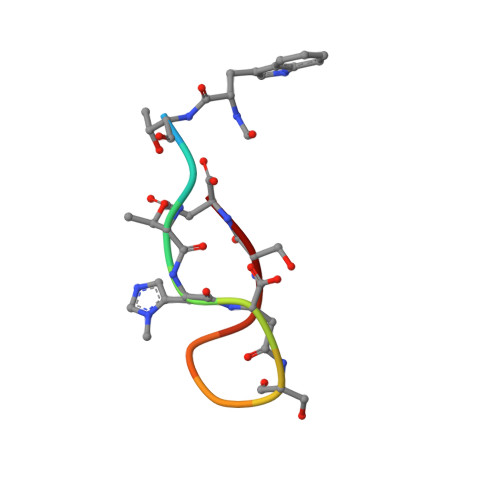
Evybactin is a DNA gyrase inhibitor that selectively kills Mycobacterium tuberculosis.
Imai, Y., Hauk, G., Quigley, J., Liang, L., Son, S., Ghiglieri, M., Gates, M.F., Morrissette, M., Shahsavari, N., Niles, S., Baldisseri, D., Honrao, C., Ma, X., Guo, J.J., Berger, J.M., Lewis, K.(2022) Nat Chem Biol 18: 1236-1244
- PubMed: 35996001
- DOI: https://doi.org/10.1038/s41589-022-01102-7
- Primary Citation of Related Structures:
7UGW - PubMed Abstract:
The antimicrobial resistance crisis requires the introduction of novel antibiotics. The use of conventional broad-spectrum compounds selects for resistance in off-target pathogens and harms the microbiome. This is especially true for Mycobacterium tuberculosis, where treatment requires a 6-month course of antibiotics. Here we show that a novel antimicrobial from Photorhabdus noenieputensis, which we named evybactin, is a potent and selective antibiotic acting against M. tuberculosis. Evybactin targets DNA gyrase and binds to a site overlapping with synthetic thiophene poisons. Given the conserved nature of DNA gyrase, the observed selectivity against M. tuberculosis is puzzling. We found that evybactin is smuggled into the cell by a promiscuous transporter of hydrophilic compounds, BacA. Evybactin is the first, but likely not the only, antimicrobial compound found to employ this unusual mechanism of selectivity.
- Antimicrobial Discovery Center, Department of Biology, Northeastern University, Boston, MA, USA.
Organizational Affiliation: