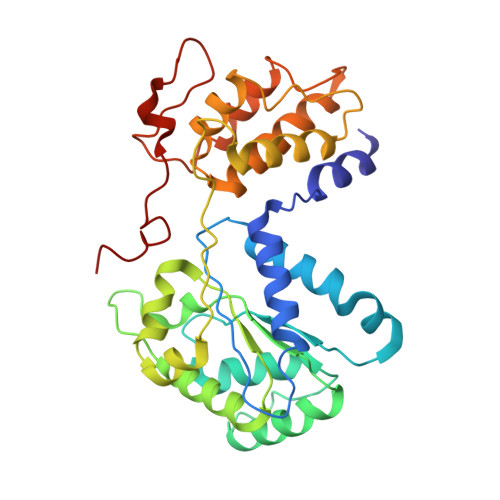
Selective TnsC recruitment enhances the fidelity of RNA-guided transposition.
Hoffmann, F.T., Kim, M., Beh, L.Y., Wang, J., Vo, P.L.H., Gelsinger, D.R., George, J.T., Acree, C., Mohabir, J.T., Fernandez, I.S., Sternberg, S.H.(2022) Nature 609: 384-393
- PubMed: 36002573
- DOI: https://doi.org/10.1038/s41586-022-05059-4
- Primary Citation of Related Structures:
7RZY, 7UFI, 7UFM - PubMed Abstract:
Bacterial transposons are pervasive mobile genetic elements that use distinct DNA-binding proteins for horizontal transmission. For example, Escherichia coli Tn7 homes to a specific attachment site using TnsD 1 , whereas CRISPR-associated transposons use type I or type V Cas effectors to insert downstream of target sites specified by guide RNAs 2,3 . Despite this targeting diversity, transposition invariably requires TnsB, a DDE-family transposase that catalyses DNA excision and insertion, and TnsC, a AAA+ ATPase that is thought to communicate between transposase and targeting proteins 4 . How TnsC mediates this communication and thereby regulates transposition fidelity has remained unclear. Here we use chromatin immunoprecipitation with sequencing to monitor in vivo formation of the type I-F RNA-guided transpososome, enabling us to resolve distinct protein recruitment events before integration. DNA targeting by the TniQ-Cascade complex is surprisingly promiscuous-hundreds of genomic off-target sites are sampled, but only a subset of those sites is licensed for TnsC and TnsB recruitment, revealing a crucial proofreading checkpoint. To advance the mechanistic understanding of interactions responsible for transpososome assembly, we determined structures of TnsC using cryogenic electron microscopy and found that ATP binding drives the formation of heptameric rings that thread DNA through the central pore, thereby positioning the substrate for downstream integration. Collectively, our results highlight the molecular specificity imparted by consecutive factor binding to genomic target sites during RNA-guided transposition, and provide a structural roadmap to guide future engineering efforts.
- Department of Biochemistry and Molecular Biophysics, Columbia University, New York, NY, USA.
Organizational Affiliation: