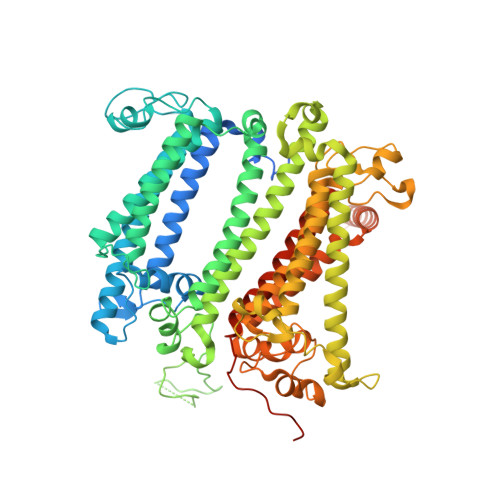
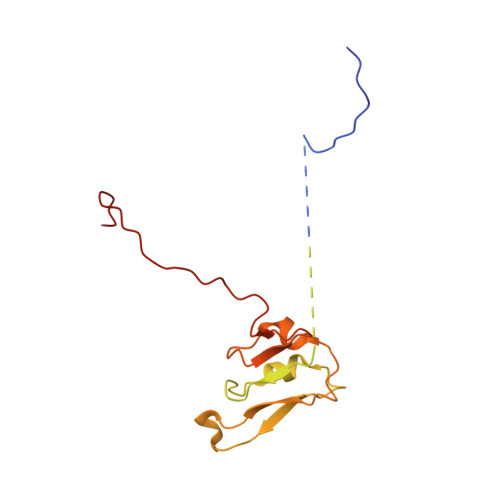
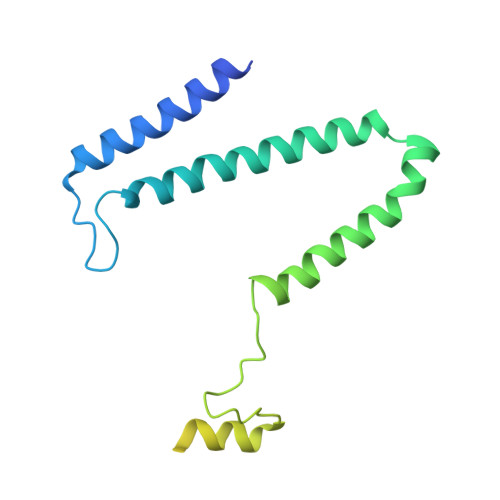
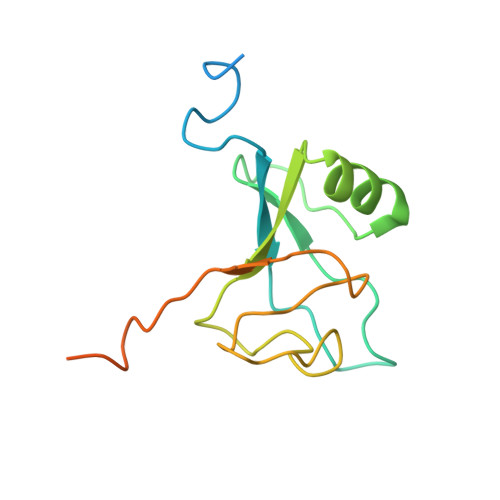
Molecular asymmetry of a photosynthetic supercomplex from green sulfur bacteria.
Puskar, R., Du Truong, C., Swain, K., Chowdhury, S., Chan, K.Y., Li, S., Cheng, K.W., Wang, T.Y., Poh, Y.P., Mazor, Y., Liu, H., Chou, T.F., Nannenga, B.L., Chiu, P.L.(2022) Nat Commun 13: 5824-5824
- PubMed: 36192412
- DOI: https://doi.org/10.1038/s41467-022-33505-4
- Primary Citation of Related Structures:
7UEA, 7UEB - PubMed Abstract:
The photochemical reaction center (RC) features a dimeric architecture for charge separation across the membrane. In green sulfur bacteria (GSB), the trimeric Fenna-Matthews-Olson (FMO) complex mediates the transfer of light energy from the chlorosome antenna complex to the RC. Here we determine the structure of the photosynthetic supercomplex from the GSB Chlorobaculum tepidum using single-particle cryogenic electron microscopy (cryo-EM) and identify the cytochrome c subunit (PscC), two accessory protein subunits (PscE and PscF), a second FMO trimeric complex, and a linker pigment between FMO and the RC core. The protein subunits that are assembled with the symmetric RC core generate an asymmetric photosynthetic supercomplex. One linker bacteriochlorophyll (BChl) is located in one of the two FMO-PscA interfaces, leading to differential efficiencies of the two energy transfer branches. The two FMO trimeric complexes establish two different binding interfaces with the RC cytoplasmic surface, driven by the associated accessory subunits. This structure of the GSB photosynthetic supercomplex provides mechanistic insight into the light excitation energy transfer routes and a possible evolutionary transition intermediate of the bacterial photosynthetic supercomplex from the primitive homodimeric RC.
- School of Molecular Sciences, Arizona State University, Tempe, AZ, 85287, USA.
Organizational Affiliation: