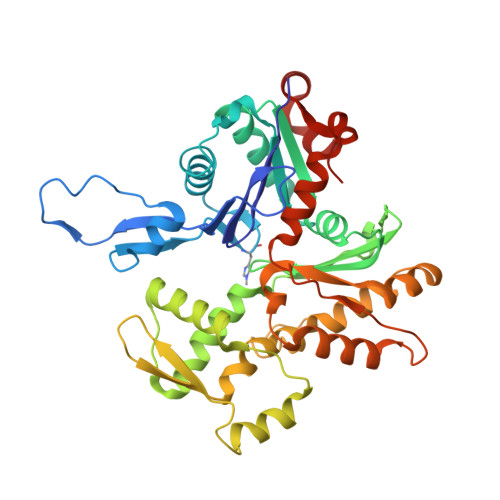
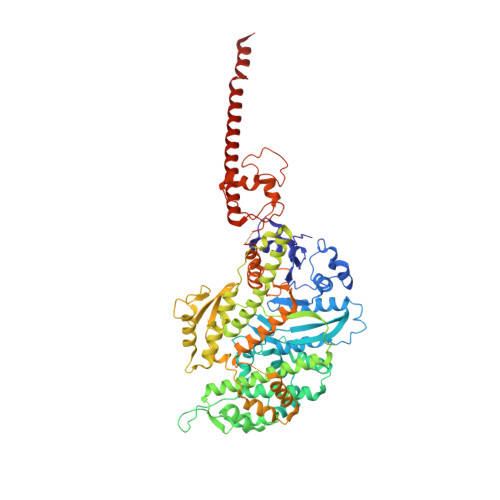
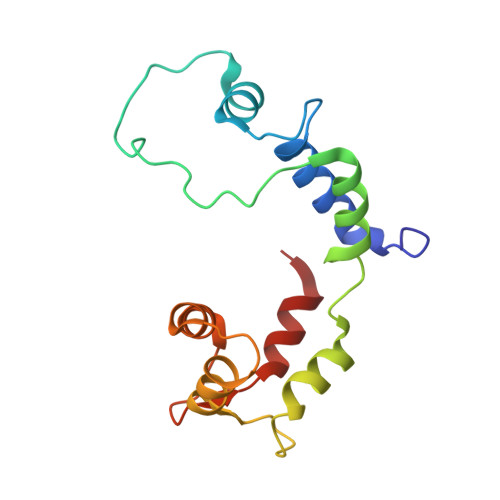
Structural basis for tunable control of actin dynamics by myosin-15 in mechanosensory stereocilia.
Gong, R., Jiang, F., Moreland, Z.G., Reynolds, M.J., de Los Reyes, S.E., Gurel, P., Shams, A., Heidings, J.B., Bowl, M.R., Bird, J.E., Alushin, G.M.(2022) Sci Adv 8: eabl4733-eabl4733
- PubMed: 35857845
- DOI: https://doi.org/10.1126/sciadv.abl4733
- Primary Citation of Related Structures:
7R8V, 7R91, 7RB8, 7RB9, 7UDT, 7UDU - PubMed Abstract:
The motor protein myosin-15 is necessary for the development and maintenance of mechanosensory stereocilia, and mutations in myosin-15 cause hereditary deafness. In addition to transporting actin regulatory machinery to stereocilia tips, myosin-15 directly nucleates actin filament ("F-actin") assembly, which is disrupted by a progressive hearing loss mutation (p.D1647G, " jordan "). Here, we present cryo-electron microscopy structures of myosin-15 bound to F-actin, providing a framework for interpreting the impacts of deafness mutations on motor activity and actin nucleation. Rigor myosin-15 evokes conformational changes in F-actin yet maintains flexibility in actin's D-loop, which mediates inter-subunit contacts, while the jordan mutant locks the D-loop in a single conformation. Adenosine diphosphate-bound myosin-15 also locks the D-loop, which correspondingly blunts actin-polymerization stimulation. We propose myosin-15 enhances polymerization by bridging actin protomers, regulating nucleation efficiency by modulating actin's structural plasticity in a myosin nucleotide state-dependent manner. This tunable regulation of actin polymerization could be harnessed to precisely control stereocilium height.
- Laboratory of Structural Biophysics and Mechanobiology, The Rockefeller University, New York, NY, USA.
Organizational Affiliation: