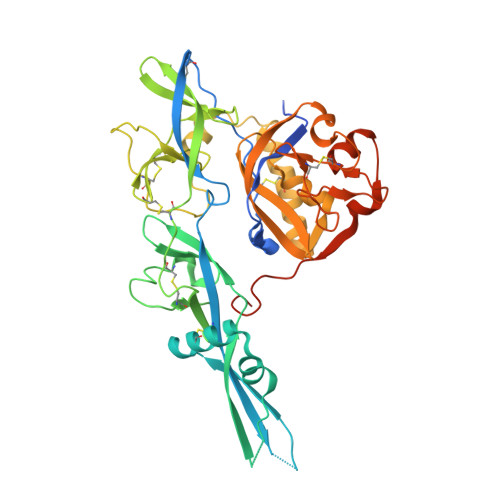
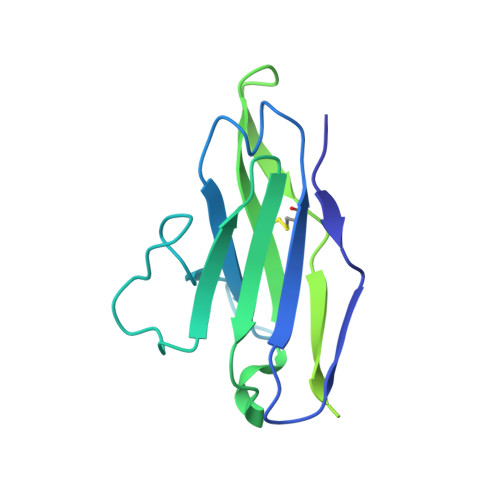
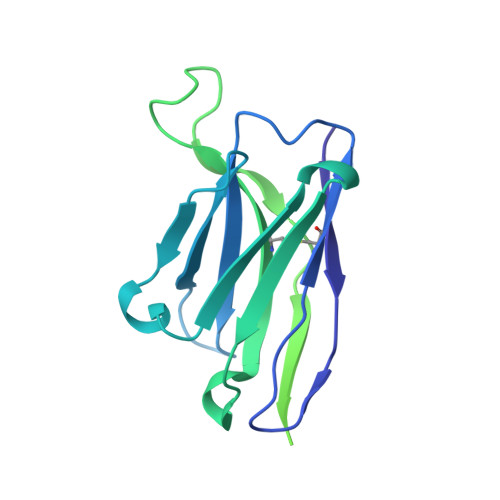
Structure of the rabies virus glycoprotein trimer bound to a prefusion-specific neutralizing antibody.
Callaway, H.M., Zyla, D., Larrous, F., de Melo, G.D., Hastie, K.M., Avalos, R.D., Agarwal, A., Corti, D., Bourhy, H., Saphire, E.O.(2022) Sci Adv 8: eabp9151-eabp9151
- PubMed: 35714192
- DOI: https://doi.org/10.1126/sciadv.abp9151
- Primary Citation of Related Structures:
7U9G - PubMed Abstract:
Rabies infection is nearly 100% lethal if untreated and kills more than 50,000 people annually, many of them children. Existing rabies vaccines target the rabies virus glycoprotein (RABV-G) but generate short-lived immune responses, likely because the protein is heterogeneous under physiological conditions. Here, we report the 3.39 Å cryo-electron microscopy structure of trimeric, prefusion RABV-G complexed with RVA122, a potently neutralizing human antibody. RVA122 binds to a quaternary epitope at the top of RABV-G, bridging domains and stabilizing RABV-G protomers in a prefusion state. RABV-G trimerization involves side-to-side interactions between the central α helix and adjacent loops, rather than contacts between central helices, and interactions among the fusion loops at the glycoprotein base. These results provide a basis from which to develop improved rabies vaccines based on RABV-G stabilized in the prefusion conformation.
- Center for Infectious Disease and Vaccine Research, La Jolla Institute for Immunology, La Jolla, CA, USA.
Organizational Affiliation: