Design and synthesis of unprecedented 9- and 10-membered cyclonucleosides with PRMT5 inhibitory activity.
Kawamura, S., Palte, R.L., Kim, H.Y., Sauri, J., Sondey, C., Mansueto, M.S., Altman, M.D., Machacek, M.R.(2022) Bioorg Med Chem 66: 116820-116820
- PubMed: 35594650
- DOI: https://doi.org/10.1016/j.bmc.2022.116820
- Primary Citation of Related Structures:
7U30 - PubMed Abstract:
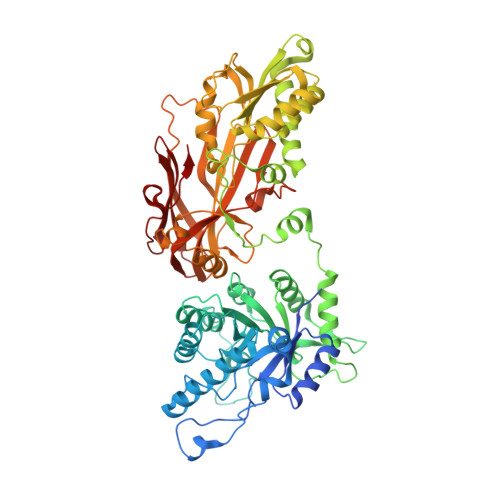
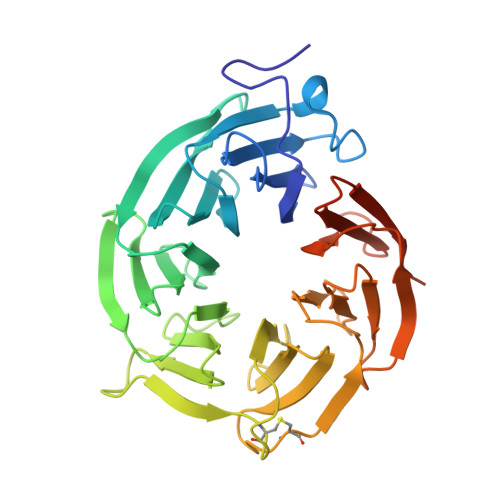
Synthesis of medium-sized rings is known to be challenging due to high transannular strain especially for 9- and 10-membered rings. Herein we report design and synthesis of unprecedented 9- and 10-membered purine 8,5'-cyclonucleosides as the first cyclonucleoside PRMT5 inhibitors. The cocrystal structure of PRMT5:MEP50 in complex with the synthesized 9-membered cyclonucleoside 1 revealed its binding mode in the SAM binding pocket of PRMT5.
- Discovery Chemistry, Merck & Co., Inc., Boston, MA 02115, United States. Electronic address: shuhei.kawamura@merck.com.
Organizational Affiliation: