A cryptic third active site in cyanophycin synthetase creates primers for polymerization
Sharon, I., Pinus, S., Grogg, M., Moitessier, N., Hilvert, D., Schmeing, T.M.(2022) Nat Commun 13: 3923
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
(2022) Nat Commun 13: 3923
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cyanophycin synthase | 879 | Synechocystis sp. PCC 6714 | Mutation(s): 0 Gene Names: cphA, D082_30240 EC: 6.3.2.29 (PDB Primary Data), 6.3.2.30 (PDB Primary Data) | 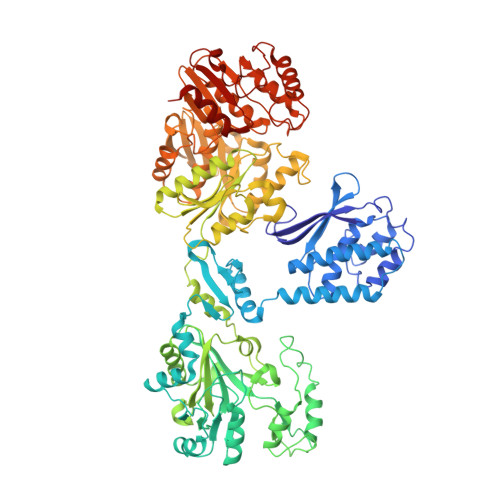 | |
UniProt | |||||
Find proteins for A0ACD6B8R9 (Synechocystis sp. (strain PCC 6714)) Explore A0ACD6B8R9 Go to UniProtKB: A0ACD6B8R9 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0ACD6B8R9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 16x(Asp-Arg) | 16 | synthetic construct | Mutation(s): 0 |  | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ATP Query on ATP | DA [auth C] EA [auth C] KA [auth D] LA [auth D] Q [auth A] | ADENOSINE-5'-TRIPHOSPHATE C10 H16 N5 O13 P3 ZKHQWZAMYRWXGA-KQYNXXCUSA-N |  | ||
| ZN Query on ZN | CA [auth C], JA [auth D], P [auth A], V [auth B] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| MG Query on MG | AA [auth C] BA [auth C] FA [auth C] GA [auth D] HA [auth D] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| 7ID Query on 7ID | E, F, G, H, I E, F, G, H, I, J, K, L | L-PEPTIDE LINKING | C10 H19 N5 O5 |  | ASP |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Canadian Institutes of Health Research (CIHR) | Canada | 178084 |