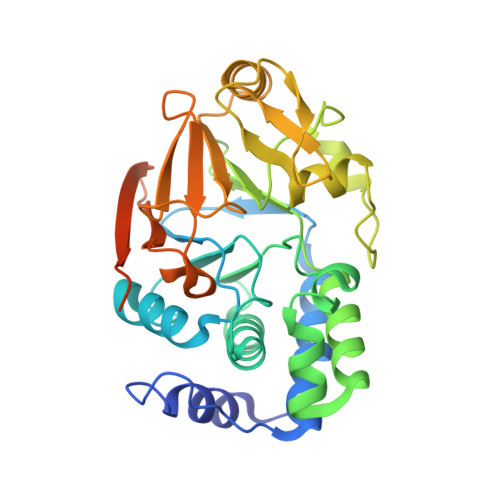
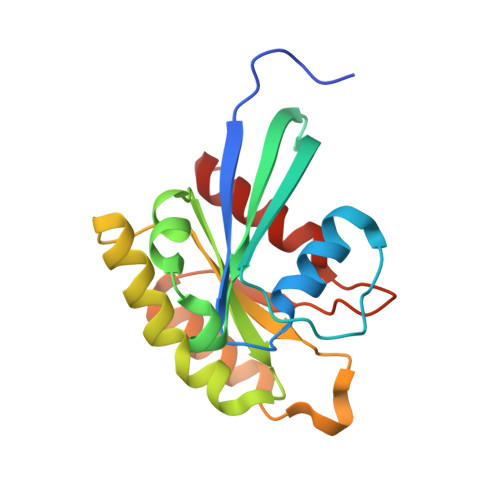
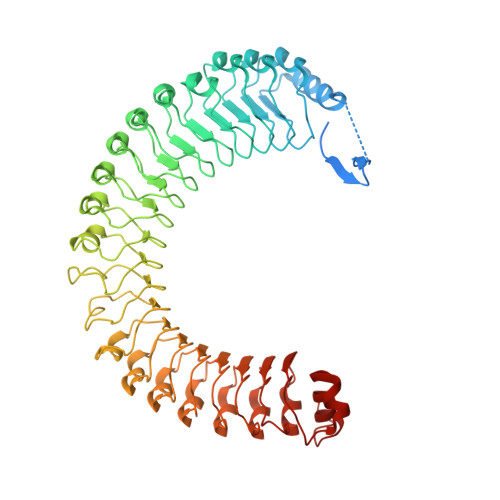
Structure of the SHOC2-MRAS-PP1C complex provides insights into RAF activation and Noonan syndrome.
Bonsor, D.A., Alexander, P., Snead, K., Hartig, N., Drew, M., Messing, S., Finci, L.I., Nissley, D.V., McCormick, F., Esposito, D., Rodriguez-Viciana, P., Stephen, A.G., Simanshu, D.K.(2022) Nat Struct Mol Biol 29: 966-977
- PubMed: 36175670
- DOI: https://doi.org/10.1038/s41594-022-00841-4
- Primary Citation of Related Structures:
7TVF, 7TVG - PubMed Abstract:
SHOC2 acts as a strong synthetic lethal interactor with MEK inhibitors in multiple KRAS cancer cell lines. SHOC2 forms a heterotrimeric complex with MRAS and PP1C that is essential for regulating RAF and MAPK-pathway activation by dephosphorylating a specific phosphoserine on RAF kinases. Here we present the high-resolution crystal structure of the SHOC2-MRAS-PP1C (SMP) complex and apo-SHOC2. Our structures reveal that SHOC2, MRAS, and PP1C form a stable ternary complex in which all three proteins synergistically interact with each other. Our results show that dephosphorylation of RAF substrates by PP1C is enhanced upon interacting with SHOC2 and MRAS. The SMP complex forms only when MRAS is in an active state and is dependent on SHOC2 functioning as a scaffolding protein in the complex by bringing PP1C and MRAS together. Our results provide structural insights into the role of the SMP complex in RAF activation and how mutations found in Noonan syndrome enhance complex formation, and reveal new avenues for therapeutic interventions.
- NCI RAS Initiative, Cancer Research Technology Program, Frederick National Laboratory for Cancer Research, Frederick, MD, USA.
Organizational Affiliation: