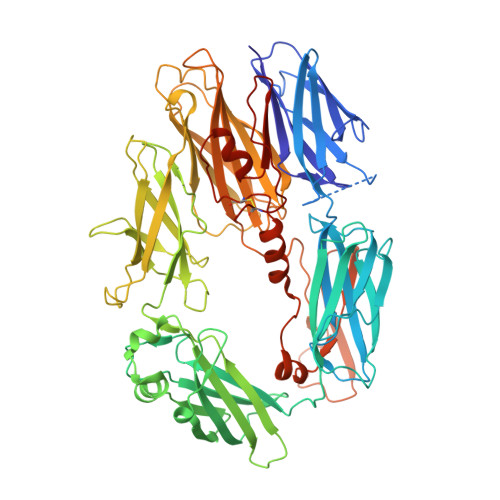
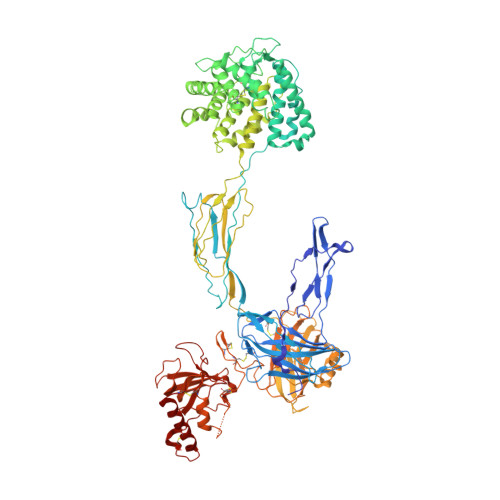
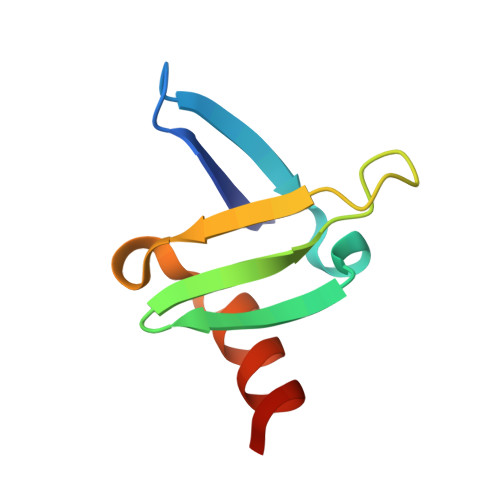
Discovery of APL-1030, a Novel, High-Affinity Nanofitin Inhibitor of C3-Mediated Complement Activation.
Garlich, J., Cinier, M., Chevrel, A., Perrocheau, A., Eyerman, D.J., Orme, M., Kitten, O., Scheibler, L.(2022) Biomolecules 12
- PubMed: 35327625
- DOI: https://doi.org/10.3390/biom12030432
- Primary Citation of Related Structures:
7TV9 - PubMed Abstract:
Uncontrolled complement activation contributes to multiple immune pathologies. Although synthetic compstatin derivatives targeting C3 and C3b are robust inhibitors of complement activation, their physicochemical and molecular properties may limit access to specific organs, development of bifunctional moieties, and therapeutic applications requiring transgenic expression. Complement-targeting therapeutics containing only natural amino acids could enable multifunctional pharmacology, gene therapies, and targeted delivery for underserved diseases. A Nanofitin library of hyperthermophilic protein scaffolds was screened using ribosome display for C3/C3b-targeting clones mimicking compstatin pharmacology. APL-1030, a recombinant 64-residue Nanofitin, emerged as the lead candidate. APL-1030 is thermostable, binds C3 (K D , 1.59 nM) and C3b (K D , 1.11 nM), and inhibits complement activation via classical (IC 50 = 110.8 nM) and alternative (IC 50 = 291.3 nM) pathways in Wieslab assays. Pharmacologic activity (determined by alternative pathway inhibition) was limited to primate species of tested sera. C3b-binding sites of APL-1030 and compstatin were shown to overlap by X-ray crystallography of C3b-bound APL-1030. APL-1030 is a novel, high-affinity inhibitor of primate C3-mediated complement activation developed from natural amino acids on the hyperthermophilic Nanofitin platform. Its properties may support novel drug candidates, enabling bifunctional moieties, gene therapy, and tissue-targeted C3 pharmacologics for diseases with high unmet need.
- Apellis Pharmaceuticals, Waltham, MA 02451, USA.
Organizational Affiliation: