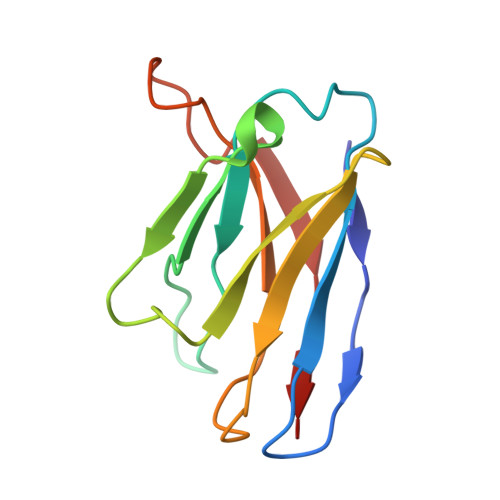
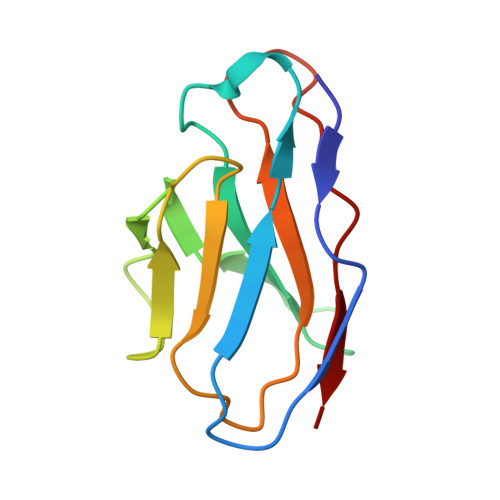
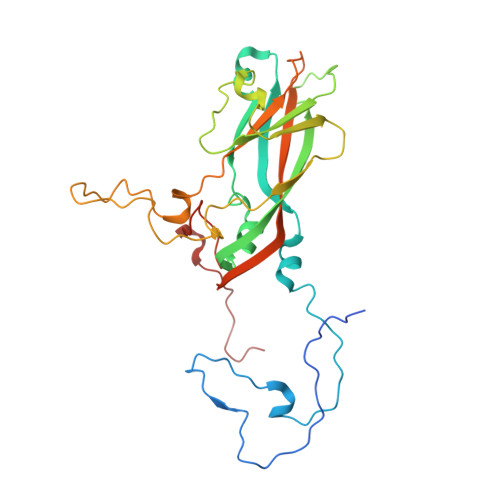
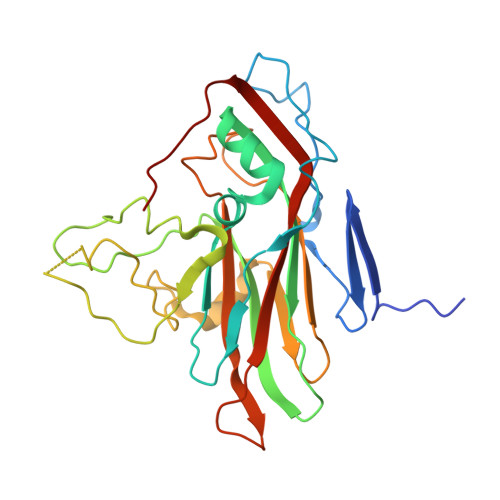
High-resolution structural analysis of enterovirus-reactive polyclonal antibodies in complex with whole virions.
Antanasijevic, A., Schulze, A.J., Reddy, V.S., Ward, A.B.(2022) PNAS Nexus 1: pgac253-pgac253
- PubMed: 36712368
- DOI: https://doi.org/10.1093/pnasnexus/pgac253
- Primary Citation of Related Structures:
7TQS, 7TQT, 7TQU - PubMed Abstract:
Non-polio enteroviruses (NPEVs) cause serious illnesses in young children and neonates, including aseptic meningitis, encephalitis, and inflammatory muscle disease, among others. While over 100 serotypes have been described to date, vaccine only exists for EV-A71. Efforts toward rationally designed pan-NPEV vaccines would greatly benefit from structural biology methods for rapid and comprehensive evaluation of vaccine candidates and elicited antibody responses. Toward this goal, we introduced a cryo-electron-microscopy-based approach for structural analysis of virus- or vaccine-elicited polyclonal antibodies (pAbs) in complex with whole NPEV virions. We demonstrated the feasibility using coxsackievirus A21 and reconstructed five structurally distinct pAbs bound to the virus. The pAbs targeted two immunodominant epitopes, one overlapping with the receptor binding site. These results demonstrate that our method can be applied to map broad-spectrum polyclonal immune responses against intact virions and define potentially cross-reactive epitopes.
- Department of Integrative, Structural and Computational Biology, The Scripps Research Institute, La Jolla, CA 92037, USA.
Organizational Affiliation: