Conformation Switch of Rieske ISP subunit is revealed by the Crystal Structure of Bacterial Cytochrome bc1 in Complex with Azoxystrobin
Xia, D., Esser, L., Zhou, F.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cytochrome b | 445 | Cereibacter sphaeroides | Mutation(s): 0 Membrane Entity: Yes | 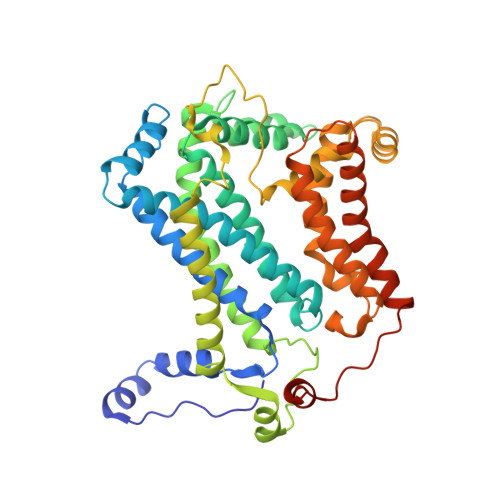 | |
UniProt | |||||
Find proteins for Q02761 (Cereibacter sphaeroides) Explore Q02761 Go to UniProtKB: Q02761 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q02761 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cytochrome c1 | 272 | Cereibacter sphaeroides | Mutation(s): 0 Gene Names: petC, fbcC Membrane Entity: Yes | 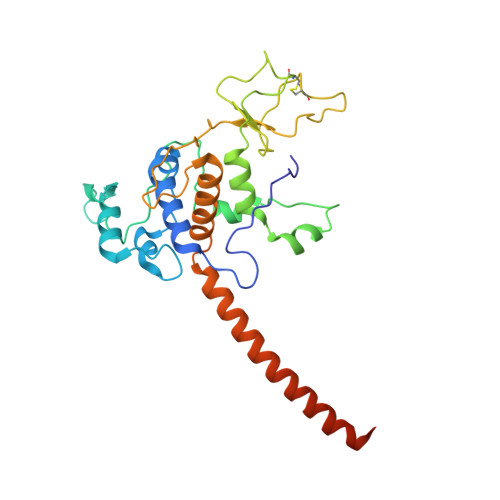 | |
UniProt | |||||
Find proteins for Q02760 (Cereibacter sphaeroides) Explore Q02760 Go to UniProtKB: Q02760 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q02760 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ubiquinol-cytochrome c reductase iron-sulfur subunit | 187 | Cereibacter sphaeroides | Mutation(s): 0 EC: 7.1.1.8 Membrane Entity: Yes | 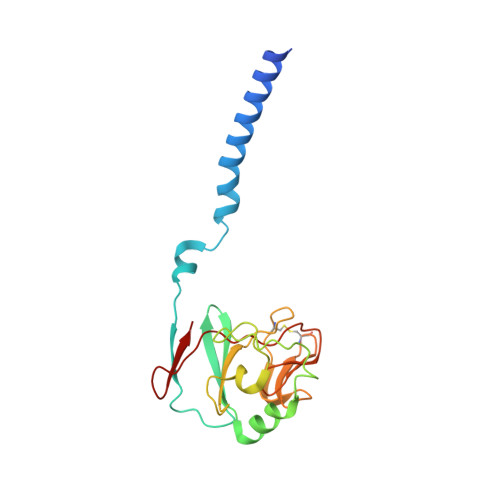 | |
UniProt | |||||
Find proteins for Q02762 (Cereibacter sphaeroides) Explore Q02762 Go to UniProtKB: Q02762 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q02762 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 14 kDa peptide of ubiquinol-cytochrome c2 oxidoreductase complex | 124 | Cereibacter sphaeroides | Mutation(s): 0 Membrane Entity: Yes |  | |
UniProt | |||||
Find proteins for P16536 (Cereibacter sphaeroides) Explore P16536 Go to UniProtKB: P16536 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P16536 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 5 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| LOP Query on LOP | L [auth A], R [auth E] | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE C35 H68 N O8 P FUUNMZKPCMPCHT-ILGKRYBBSA-N |  | ||
| HEC Query on HEC | M [auth B], S [auth F] | HEME C C34 H34 Fe N4 O4 HXQIYSLZKNYNMH-LJNAALQVSA-N |  | ||
| HEM Query on HEM | I [auth A], J [auth A], O [auth E], P [auth E] | PROTOPORPHYRIN IX CONTAINING FE C34 H32 Fe N4 O4 KABFMIBPWCXCRK-RGGAHWMASA-L |  | ||
| PQU (Subject of Investigation/LOI) Query on PQU | K [auth A], Q [auth E] | (5S)-3-anilino-5-methyl-5-(6-phenoxypyridin-3-yl)-1,3-oxazolidine-2,4-dione C21 H17 N3 O4 QOFLFGUNPBNKDO-NRFANRHFSA-N |  | ||
| FES Query on FES | N [auth C], T [auth G] | FE2/S2 (INORGANIC) CLUSTER Fe2 S2 NIXDOXVAJZFRNF-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| RECONSTRUCTION | cisTEM | 1.0.0 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Cancer Institute (NIH/NCI) | United States | -- |