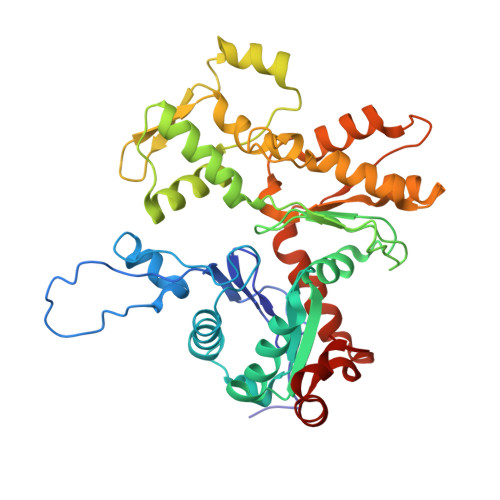
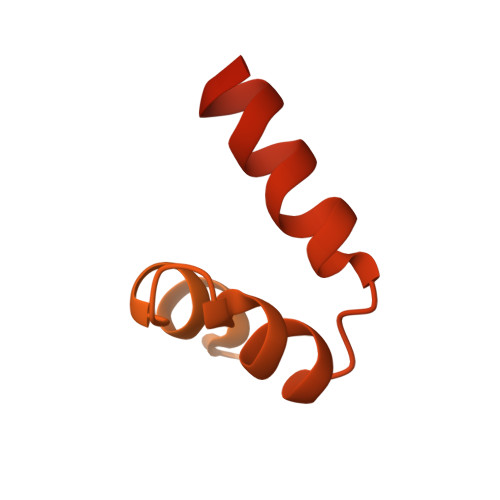
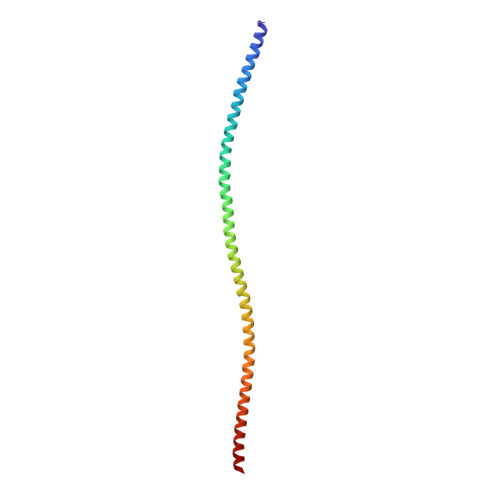Cryo-Electron Microscopy Reveals Cardiac Myosin Binding Protein-C M-Domain Interactions with the Thin Filament.
Risi, C.M., Villanueva, E., Belknap, B., Sadler, R.L., Harris, S.P., White, H.D., Galkin, V.E.(2022) J Mol Biology 434: 167879-167879
- PubMed: 36370805
- DOI: https://doi.org/10.1016/j.jmb.2022.167879
- Primary Citation of Related Structures:
7TIJ, 7TIT, 7TJ7 - PubMed Abstract:
Cardiac myosin binding protein C (cMyBP-C) modulates cardiac contraction via direct interactions with cardiac thick (myosin) and thin (actin) filaments (cTFs). While its C-terminal domains (e.g. C8-C10) anchor cMyBP-C to the backbone of the thick filament, its N-terminal domains (NTDs) (e.g. C0, C1, M, and C2) bind to both myosin and actin to accomplish its dual roles of inhibiting thick filaments and activating cTFs. While the positions of C0, C1 and C2 on cTF have been reported, the binding site of the M-domain on the surface of the cTF is unknown. Here, we used cryo-EM to reveal that the M-domain interacts with actin via helix 3 of its ordered tri-helix bundle region, while the unstructured part of the M-domain does not maintain extensive interactions with actin. We combined the recently obtained structure of the cTF with the positions of all the four NTDs on its surface to propose a complete model of the NTD binding to the cTF. The model predicts that the interactions of the NTDs with the cTF depend on the activation state of the cTF. At the peak of systole, when bound to the extensively activated cTF, NTDs would inhibit actomyosin interactions. In contrast, at falling Ca 2+ levels, NTDs would not compete with the myosin heads for binding to the cTF, but would rather promote formation of active cross-bridges at the adjacent regulatory units located at the opposite cTF strand. Our structural data provides a testable model of the cTF regulation by the cMyBP-C.
- Department of Physiological Sciences, Eastern Virginia Medical School, Norfolk, VA 23507, USA.
Organizational Affiliation: