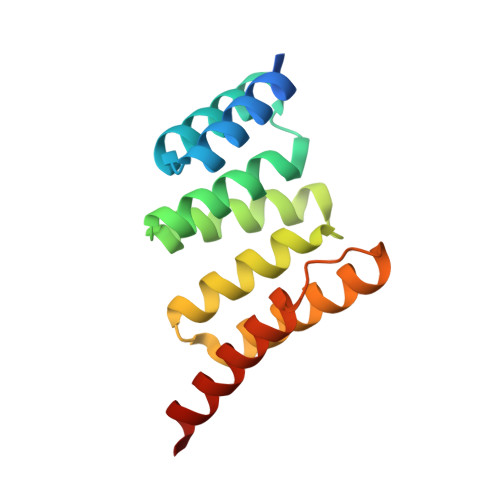
Discovery and Structure-Based Design of Macrocyclic Peptides Targeting STUB1.
Ng, S., Brueckner, A.C., Bahmanjah, S., Deng, Q., Johnston, J.M., Ge, L., Duggal, R., Habulihaz, B., Barlock, B., Ha, S., Sadruddin, A., Yeo, C., Strickland, C., Peier, A., Henry, B., Sherer, E.C., Partridge, A.W.(2022) J Med Chem
- PubMed: 35853179
- DOI: https://doi.org/10.1021/acs.jmedchem.2c00406
- Primary Citation of Related Structures:
7TB1 - PubMed Abstract:
Recent evidence suggests that deletion of STUB1─a pivotal negative regulator of interferon-γ sensing─may potentially clear malignant cells. However, current studies rely primarily on genetic approaches, as pharmacological inhibitors of STUB1 are lacking. Identifying a tool compound will be a step toward validating the target in a broader therapeutic sense. Herein, screening more than a billion macrocyclic peptides resulted in STUB1 binders, which were further optimized by a structure-enabled in silico design. The strategy to replace the macrocyclic peptides' hydrophilic and solvent-exposed region with a hydrophobic scaffold improved cellular permeability while maintaining the binding conformation. Further substitution of the permeability-limiting terminal aspartic acid with a tetrazole bioisostere retained the binding to a certain extent while improving permeability, suggesting a path forward. Although not optimal for cellular study, the current lead provides a valuable template for further development into selective tool compounds for STUB1 to enable target validation.
- Quantitative Biosciences, MSD, 8 Biomedical Grove, Singapore 138665.
Organizational Affiliation: