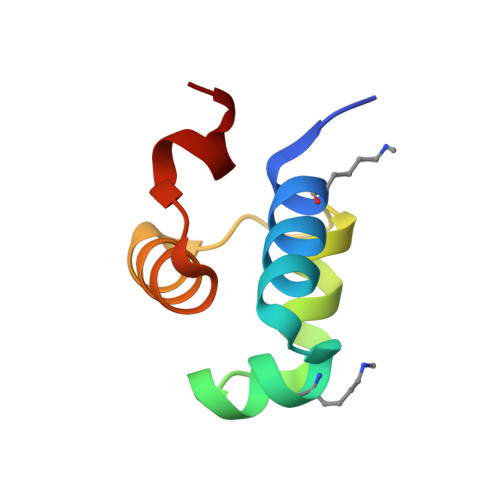
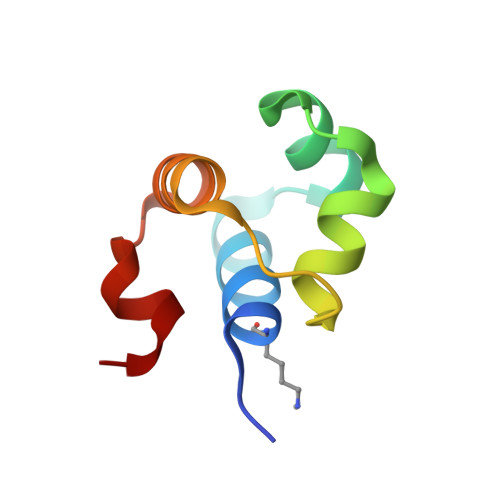
Crystal structure report of the ImmR transcriptional regulator DNA-binding domain of the Bacillus subtilis ICEBs1 transposon.
Caliandro, R., de Diego, I., Gomis-Ruth, F.X.(2022) Sci Rep 12: 5258-5258
- PubMed: 35347179
- DOI: https://doi.org/10.1038/s41598-022-09237-2
- Primary Citation of Related Structures:
7T8I - PubMed Abstract:
Bacillus subtilis is a commensal member of the human oral and gut microbiomes, which can become infectious to immunocompromised patients. It possesses a conjugative transposon, ICEBs1, which includes > 20 genes and can be passed by horizontal gene transfer to other bacteria, including pathogenic Bacillus anthracis and Listeria monocytogenes. ICEBs1 is regulated by the ImmR/ImmA tandem, which are a transcriptional repressor that constitutively blocks transcription and a metallopeptidase that acts as anti-repressor and inactivates ImmR by proteolytic cleavage. We here report the production and purification of 127-residue ImmR from ICEBs1 and the crystal structure of its DNA-binding domain. It features a five-helix bundle centred on a helix-turn-helix motif potentially binding the major grove of double-stranded target DNA. ImmR shows structural and mechanistic similarity with the B. subtilis SinR repressor, which is engaged in sporulation inhibition.
- Proteolysis Laboratory, Department of Structural Biology, Molecular Biology Institute of Barcelona (CSIC), Barcelona Science Park, C/Baldiri Reixac, 15-21, 08028, Barcelona, Catalonia, Spain.
Organizational Affiliation: